Multiplexed CRISPR gene editing in primary human islet cells with Cas9 ribonucleoprotein
- PMID: 38205242
- PMCID: PMC10777115
- DOI: 10.1016/j.isci.2023.108693
Multiplexed CRISPR gene editing in primary human islet cells with Cas9 ribonucleoprotein
Abstract
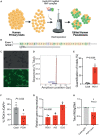
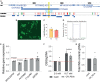
Successful genome editing in primary human islets could reveal features of the genetic regulatory landscape underlying β cell function and diabetes risk. Here, we describe a CRISPR-based strategy to interrogate functions of predicted regulatory DNA elements using electroporation of a complex of Cas9 ribonucleoprotein (Cas9 RNP) and guide RNAs into primary human islet cells. We successfully targeted coding regions including the PDX1 exon 1, and non-coding DNA linked to diabetes susceptibility. CRISPR-Cas9 RNP approaches revealed genetic targets of regulation by DNA elements containing candidate diabetes risk SNPs, including an in vivo enhancer of the MPHOSPH9 gene. CRISPR-Cas9 RNP multiplexed targeting of two cis-regulatory elements linked to diabetes risk in PCSK1, which encodes an endoprotease crucial for Insulin processing, also demonstrated efficient simultaneous editing of PCSK1 regulatory elements, resulting in impaired β cell PCSK1 regulation and Insulin secretion. Multiplex CRISPR-Cas9 RNP provides powerful approaches to investigate and elucidate human islet cell gene regulation in health and diabetes.
Keywords: Techniques in genetics; biology experimental methods; cell biology; human genetics.
© 2023 The Author(s).
Conflict of interest statement
A.M. is a co-founder of Arsenal Biosciences, Function Bio, Spotlight Therapeutics, and Survey Genomics, serves on the boards of directors at Function Bio, Spotlight Therapeutics, and Survey Genomics, is a member of the scientific advisory boards of Arsenal Biosciences, Function Bio, Spotlight Therapeutics, Survey Genomics, NewLimit, Amgen, Tenaya, and Lightcast, owns stock in Arsenal Biosciences, Function Bio, Spotlight Therapeutics, NewLimit, Survey Genomics, Tenaya, and Lightcast, and has received fees from Arsenal Biosciences, Spotlight Therapeutics, NewLimit, 23andMe, PACT Pharma, Juno Therapeutics, Tenaya, Lightcast, Trizell, Vertex, Merck, Amgen, Genentech, AlphaSights, Rupert Case Management, Bernstein, GLG, ClearView Healthcare Partners, and ALDA. A.M. is an investor in and informal advisor to Offline Ventures and a client of EPIQ. The Marson laboratory has received research support from Juno Therapeutics, Epinomics, Sanofi, GlaxoSmithKline, Gilead, and Anthem. A.L.G.’s spouse holds stock options in Roche and is an employee of Genentech.
Figures



Update of
-
Multiplexed CRISPR gene editing in primary human islet cells with Cas9 ribonucleoprotein.bioRxiv [Preprint]. 2023 Sep 17:2023.09.16.558090. doi: 10.1101/2023.09.16.558090. bioRxiv. 2023. Update in: iScience. 2023 Dec 08;27(1):108693. doi: 10.1016/j.isci.2023.108693. PMID: 37745551 Free PMC article. Updated. Preprint.
Similar articles
-
Multiplexed CRISPR gene editing in primary human islet cells with Cas9 ribonucleoprotein.bioRxiv [Preprint]. 2023 Sep 17:2023.09.16.558090. doi: 10.1101/2023.09.16.558090. bioRxiv. 2023. Update in: iScience. 2023 Dec 08;27(1):108693. doi: 10.1016/j.isci.2023.108693. PMID: 37745551 Free PMC article. Updated. Preprint.
-
Modulating binding affinity of aptamer-based loading constructs enhances extracellular vesicle-mediated CRISPR/Cas9 delivery.J Control Release. 2025 Aug 10;384:113853. doi: 10.1016/j.jconrel.2025.113853. Epub 2025 May 18. J Control Release. 2025. PMID: 40393529
-
Cell-Penetrating Peptides and CRISPR-Cas9: A Combined Strategy for Human Genetic Disease Therapy.Hum Gene Ther. 2024 Oct;35(19-20):781-797. doi: 10.1089/hum.2024.020. Hum Gene Ther. 2024. PMID: 39276086 Review.
-
CRISPR-Cas9 mediated proteinase 3 autoantigen deletion as a treatment strategy for anti-neutrophil cytoplasmic autoantibody-associated vasculitis.Kidney Int. 2025 Jul;108(1):145-149. doi: 10.1016/j.kint.2025.03.020. Epub 2025 Apr 21. Kidney Int. 2025. PMID: 40268165
-
Transferable approaches to CRISPR-Cas9 induced genome editing in non-model insects: a brief guide.Front Zool. 2025 Jul 7;22(1):13. doi: 10.1186/s12983-025-00566-2. Front Zool. 2025. PMID: 40624545 Free PMC article. Review.
Cited by
-
Harnessing beta-cell replication: advancing molecular insights to regenerative therapies in diabetes.Front Endocrinol (Lausanne). 2025 Jun 19;16:1612576. doi: 10.3389/fendo.2025.1612576. eCollection 2025. Front Endocrinol (Lausanne). 2025. PMID: 40612435 Free PMC article. Review.
-
Analysis of biased allelic enhancer activity of schizophrenia-linked common variants.Commun Biol. 2025 Jul 10;8(1):1034. doi: 10.1038/s42003-025-08456-3. Commun Biol. 2025. PMID: 40640362 Free PMC article.
-
Harnessing bacterial immunity: CRISPR-Cas system as a versatile tool in combating pathogens and revolutionizing medicine.Front Cell Infect Microbiol. 2025 May 30;15:1588446. doi: 10.3389/fcimb.2025.1588446. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40521034 Free PMC article. Review.
-
Isolation of live human δ cells for genetic and functional analysis.Mol Metab. 2025 Aug;98:102188. doi: 10.1016/j.molmet.2025.102188. Epub 2025 Jun 11. Mol Metab. 2025. PMID: 40514008 Free PMC article.
-
Predictive Prioritization of Enhancers Associated with Pancreas Disease Risk.bioRxiv [Preprint]. 2024 Sep 13:2024.09.07.611794. doi: 10.1101/2024.09.07.611794. bioRxiv. 2024. PMID: 39314336 Free PMC article. Preprint.
References
-
- Gloyn A.L., Pearson E.R., Antcliff J.F., Proks P., Bruining G.J., Slingerland A.S., Howard N., Srinivasan S., Silva J.M.C.L., Molnes J., et al. Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N. Engl. J. Med. 2004;350:1838–1849. - PubMed
-
- Njølstad P.R., Søvik O., Cuesta-Muñoz A., Bjørkhaug L., Massa O., Barbetti F., Undlien D.E., Shiota C., Magnuson M.A., Molven A., et al. Neonatal diabetes mellitus due to complete glucokinase deficiency. N. Engl. J. Med. 2001;344:1588–1592. - PubMed
-
- Stoffers D.A., Zinkin N.T., Stanojevic V., Clarke W.L., Habener J.F. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat. Genet. 1997;15:106–110. - PubMed
-
- Mahajan A., Taliun D., Thurner M., Robertson N.R., Torres J.M., Rayner N.W., Payne A.J., Steinthorsdottir V., Scott R.A., Grarup N., et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 2018;50:1505–1513. - PMC - PubMed

