Comprehensive characterization and database construction of immune repertoire in the largest Chinese glioma cohort
- PMID: 38205245
- PMCID: PMC10777385
- DOI: 10.1016/j.isci.2023.108661
Comprehensive characterization and database construction of immune repertoire in the largest Chinese glioma cohort
Abstract
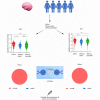
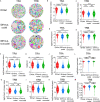
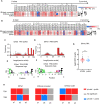
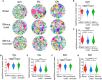
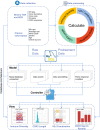
Immune receptor repertoire is valuable for developing immunotherapeutic interventions, but remains poorly understood across glioma subtypes including IDH wild type, IDH mutation without 1p/19q codeletion (IDHmut-noncodel) and IDH mutation with 1p/19q codeletion (IDHmut-codel). We assembled over 320,000 TCR/BCR clonotypes from the largest glioma cohort of 913 RNA sequencing samples in the Chinese population, finding that immune repertoire diversity was more prominent in the IDH wild type (the most aggressive glioma). Fewer clonotypes were shared within each glioma subtype, indicating high heterogeneity of the immune repertoire. The TRA-CDR3 was longer in private than in public clonotypes in IDH wild type. CDR3 variable motifs had higher proportions of hydrophobic residues in private than in public clonotypes, suggesting private CDR3 sequences have greater potential for tumor antigen recognition. Finally, we developed GTABdb, a web-based database designed for hosting, exploring, visualizing, and analyzing glioma immune repertoire. Our study will facilitate developing glioma immunotherapy.
Keywords: Health sciences; Immunology.
© 2023 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Wen P.Y., Weller M., Lee E.Q., Alexander B.M., Barnholtz-Sloan J.S., Barthel F.P., Batchelor T.T., Bindra R.S., Chang S.M., Chiocca E.A., et al. Glioblastoma in adults: a Society for Neuro-Oncology (SNO) and European Society of Neuro-Oncology (EANO) consensus review on current management and future directions. Neuro Oncol. 2020;22:1073–1113. - PMC - PubMed
-
- Louis D.N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W.K., Ohgaki H., Wiestler O.D., Kleihues P., Ellison D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–820. - PubMed
Associated data
LinkOut - more resources
Full Text Sources

