Identification of differentially expressed genes, signaling pathways and immune infiltration in postmenopausal osteoporosis by integrated bioinformatics analysis
- PMID: 38205281
- PMCID: PMC10777010
- DOI: 10.1016/j.heliyon.2023.e23794
Identification of differentially expressed genes, signaling pathways and immune infiltration in postmenopausal osteoporosis by integrated bioinformatics analysis
Abstract
Background: Postmenopausal osteoporosis is a systemic metabolic disorder typified by an imbalance in bone turnover, where bone resorption supersedes bone formation. This imbalance primarily arises from a decline in bone mass induced by estrogen deficiency, and an elevated risk of fractures resulting from degradation of bone microstructure. Despite recognizing these changes, the precise causative factors and potential molecular pathways remain elusive. In this study, we aimed to identify differentially expressed genes (DEGs), associated pathways, and the role of immune infiltration in osteoporosis, leveraging an integrated bioinformatics approach to shed light on potential underlying molecular mechanisms.
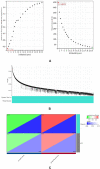
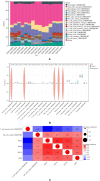
Methods: We retrieved the expression profiles of GSE230665 from the Gene Expression Omnibus database, comprising 15 femur samples, including 12 postmenopausal osteoporosis samples and 3 normal controls. From the aggregated microarray datasets, we derived differentially expressed genes (DEGs) for further bioinformatics analysis. We used WGCNA, analyzed DEGs, PPI, and conducted GO analysis to identify pivotal genes. We then used the CIBERSORT method to explore the degree of immune cell infiltration within femur specimens affected by postmenopausal osteoporosis. To probe into the relationship between pivotal genes and infiltrating immune cells, we conducted correlation analysis.
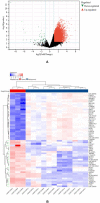
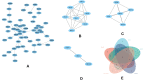
Results: We identified a total of 12,204 DEGs. Among these, 12,157 were up-regulated, and 47 were down-regulated. GO and KEGG pathway analyses indicated that these DEGs predominantly targeted cellular protein localization activity and associated signaling pathways. The protein-protein interaction network highlighted four central hub-genes: RPL31, RPL34, EEF1G, and BPTF. Principal component analysis indicated a positive correlation between the expression of these genes and resting NK cells (as per CIBERSORT). In contrast, the expression of RPL31, RPL34, and EEF1G showed a negative correlation with T cells (gamma delta per CIBERSORT).
Conclusions: Immune infiltration plays a role in the development of osteoporosis.
Keywords: Bioinformatics; Hub genes; Postmenopausal osteoporosis; WGCNA.
© 2023 The Authors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Yu F., Xia W. The epidemiology of osteoporosis, associated fragility fractures, and management gap in China. Arch. Osteoporosis. 2019;14(1):32. - PubMed
-
- Cipriani C., Pepe J., Bertoldo F., Bianchi G., Cantatore F.P., Corrado A., Di Stefano M., Frediani B., Gatti D., Giustina A., Porcelli T., Isaia G., Rossini M., Nieddu L., Minisola S., Girasole G., Pedrazzoni M. The epidemiology of osteoporosis in Italian postmenopausal women according to the National Bone Health Alliance (NBHA) diagnostic criteria: a multicenter cohort study. Journal of endocrinological investigation. 2018;41(4):431–438. - PubMed
-
- Riggs B.L., Khosla S., Melton L.J. A unitary model for involutional osteoporosis: estrogen deficiency causes both type I and type II osteoporosis in postmenopausal women and contributes to bone loss in aging men. J. Bone Miner. Res. : the official journal of the American Society for Bone and Mineral Research. 1998;13(5):763–773. - PubMed
-
- Fujita T. Calcium intake, calcium absorption, and osteoporosis. Calcif. Tissue Int. 1996;58(4):215. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

