Epigenetic variation mediated by lncRNAs accounts for adaptive genomic differentiation of the endemic blue mussel Mytiluschilensis
- PMID: 38205306
- PMCID: PMC10776947
- DOI: 10.1016/j.heliyon.2023.e23695
Epigenetic variation mediated by lncRNAs accounts for adaptive genomic differentiation of the endemic blue mussel Mytiluschilensis
Abstract
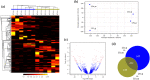
Epigenetic variation affects gene expression without altering the underlying DNA sequence of genes controlling ecologically relevant phenotypes through different mechanisms, one of which is long non-coding RNAs (lncRNAs). This study identified and evaluated the gene expression of lncRNAs in the gill and mantle tissues of Mytilus chilensis individuals from two ecologically different sites: Cochamó (41°S) and Yaldad (43°S), southern Chile, both impacted by climatic-related conditions and by mussel farming given their use as seedbeds. Sequences identified as lncRNAs exhibited tissue-specific differences, mapping to 3.54 % of the gill transcriptome and 1.96 % of the mantle transcriptome, representing an average of 2.76 % of the whole transcriptome. Using a high fold change value (≥|100|), we identified 43 and 47 differentially expressed lncRNAs (DE-lncRNAs) in the gill and mantle tissue of individuals sampled from Cochamó and 21 and 17 in the gill and mantle tissue of individuals sampled from Yaldad. Location-specific DE-lncRNAs were also detected in Cochamó (65) and Yaldad (94) samples. Via analysis of the differential expression of neighboring protein-coding genes, we identified enriched GO terms related to metabolic, genetic, and environmental information processing and immune system functions, reflecting how the impact of local ecological conditions may influence the M. chilensis (epi)genome expression. These DE-lncRNAs represent complementary biomarkers to DNA sequence variation for maintaining adaptive differences and phenotypic plasticity to cope with natural and human-driven perturbations.
Keywords: Differential gene expression; Epigenetics; Genome functioning; Mytilus chilensis; lncRNAs.
© 2023 The Authors. Published by Elsevier Ltd.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



Similar articles
-
Decoding Local Adaptation in the Exploited Native Marine Mussel Mytilus chilensis: Genomic Evidence from a Reciprocal Transplant Experiment.Int J Mol Sci. 2025 Jan 23;26(3):931. doi: 10.3390/ijms26030931. Int J Mol Sci. 2025. PMID: 39940706 Free PMC article.
-
Adaptive Differences in Gene Expression in Farm-Impacted Seedbeds of the Native Blue Mussel Mytilus chilensis.Front Genet. 2021 May 20;12:666539. doi: 10.3389/fgene.2021.666539. eCollection 2021. Front Genet. 2021. PMID: 34093658 Free PMC article.
-
Adaptive mitochondrial genome functioning in ecologically different farm-impacted natural seedbeds of the endemic blue mussel Mytilus chilensis.Comp Biochem Physiol Part D Genomics Proteomics. 2022 Jun;42:100955. doi: 10.1016/j.cbd.2021.100955. Epub 2022 Jan 4. Comp Biochem Physiol Part D Genomics Proteomics. 2022. PMID: 35065314
-
Epigenomics in stress tolerance of plants under the climate change.Mol Biol Rep. 2023 Jul;50(7):6201-6216. doi: 10.1007/s11033-023-08539-6. Epub 2023 Jun 9. Mol Biol Rep. 2023. PMID: 37294468 Review.
-
Long Non-Coding RNAs As Epigenetic Regulators in Cancer.Curr Pharm Des. 2019;25(33):3563-3577. doi: 10.2174/1381612825666190830161528. Curr Pharm Des. 2019. PMID: 31470781 Review.
Cited by
-
Decoding Local Adaptation in the Exploited Native Marine Mussel Mytilus chilensis: Genomic Evidence from a Reciprocal Transplant Experiment.Int J Mol Sci. 2025 Jan 23;26(3):931. doi: 10.3390/ijms26030931. Int J Mol Sci. 2025. PMID: 39940706 Free PMC article.
References
-
- Hofmann G.E. Ecological epigenetics in marine metazoans. Front. Mar. Sci. 2017;4:4. doi: 10.3389/fmars.2017.00004. - DOI
-
- Lamka G.F., Harder A.M., Sundaram M., Schwartz T.S., Christie M.R., DeWoody J.A., Willoughby J.R. Epigenetics in ecology, evolution, and conservation. Front. Ecol. Evol. 2022;10 doi: 10.3389/fevo.2022.871791. - DOI
LinkOut - more resources
Full Text Sources

