Potent Suppression of Prostate Cancer Cell Growth and Eradication of Cancer Stem Cells by CD44-targeted Nanoliposome-quercetin Nanoparticles
- PMID: 38205358
- PMCID: PMC10774486
- DOI: 10.15430/JCP.2023.28.4.160
Potent Suppression of Prostate Cancer Cell Growth and Eradication of Cancer Stem Cells by CD44-targeted Nanoliposome-quercetin Nanoparticles
Abstract
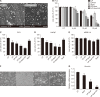
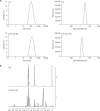
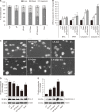
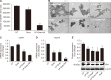
The bioavailability of quercetin, a natural compound, is hindered by low solubility, limited absorption, and restricted systemic availability. Therefore, encapsulating it in biocompatible nanoparticles presents a promising solution. This study aimed to target prostate cancer stem cells (CSCs) overexpressing CD44+ receptors as well as cancer cells, employing quercetin-loaded hyaluronic acid-modified nanoliposomes (LP-Quer-HA). Synthesized via a green ethanol injection method, these nanoliposomes had an average diameter of 134 nm and an impressive loading efficiency of 96.9%. Human prostate cancer cells were treated with either 10 μM of free quercetin or the same concentration delivered by LP-Quer-HA for 72 hours. Free quercetin reduced androgen-resistant PC3 cell viability by 16%, while LP-Quer-HA significantly increased cell death to 60%. It induced apoptosis, upregulating cytochrome c, Bax, caspases 3 and 8, and downregulating survivin and Bcl-2 expression. Compared to free quercetin, LP-Quer-HA upregulated E-cadherin expression while inhibiting cell migration and reducing the expression of fibronectin, N-cadherin, and MMP9. Treatment of PC3 cell tumor spheroids with LP-Quer-HA decreased the number of CD44 cells and expression of CD44, Oct3/4 and Wnt. Moreover, LP-Quer-HA inhibited p-ERK expression while increasing p38/MAPK and NF-κB protein expression. In androgen-sensitive LNCaP cells, LP-Quer-HA efficacy was notable, reducing cell viability from 10% to 52% compared to free quercetin. Utilizing HA-modified nanoliposomes as a quercetin delivery system enhanced its potency at lower concentrations, reducing the CD44+ cell population and effectively impeding prostate cancer cell proliferation and migration. These findings underscore the potential of quercetin-loaded cationic nanoliposomes as a robust therapeutic approach.
Keywords: Liposomes; Nanoparticles; Neoplasms; Prostate cancer; Quercetin; Therapeutic use.
Copyright © 2023 Korean Society of Cancer Prevention.
Conflict of interest statement
CONFLICTS OF INTEREST No potential conflicts of interest were disclosed.
Figures


References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
