Control of successive unequal cell divisions by neural cell fate regulators determines embryonic neuroblast cell size
- PMID: 38205939
- PMCID: PMC10911278
- DOI: 10.1242/dev.200981
Control of successive unequal cell divisions by neural cell fate regulators determines embryonic neuroblast cell size
Abstract
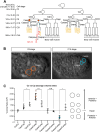
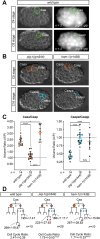
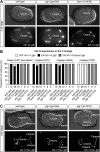
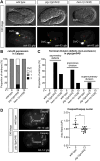
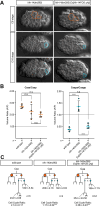
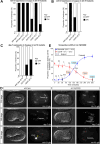
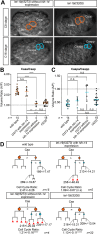
Asymmetric cell divisions often generate daughter cells of unequal size in addition to different fates. In some contexts, daughter cell size asymmetry is thought to be a key input to specific binary cell fate decisions. An alternative possibility is that unequal division is a mechanism by which a variety of cells of different sizes are generated during embryonic development. We show here that two unequal cell divisions precede neuroblast formation in the C lineage of Caenorhabditis elegans. The equalisation of these divisions in a pig-1/MELK mutant background has little effect on neuroblast specification. Instead, we demonstrate that let-19/MDT13 is a regulator of the proneural basic helix-loop-helix transcription factor hlh-14/ASCL1 and find that both are required to concomitantly regulate the acquisition of neuroblast identity and neuroblast cell size. Thus, embryonic neuroblast cell size in this lineage is progressively regulated in parallel with identity by key neural cell fate regulators. We propose that key cell fate determinants have a previously unappreciated function in regulating unequal cleavage, and therefore cell size, of the progenitor cells whose daughter cell fates they then go on to specify.
Keywords: C. elegans; Asymmetric division; Neuronal specification; Proneural.
© 2024. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures
Similar articles
-
Caenorhabditis elegans PIG-1/MELK acts in a conserved PAR-4/LKB1 polarity pathway to promote asymmetric neuroblast divisions.Genetics. 2013 Mar;193(3):897-909. doi: 10.1534/genetics.112.148106. Epub 2012 Dec 24. Genetics. 2013. PMID: 23267054 Free PMC article.
-
Caenorhabditis elegans CES-1 Snail Represses pig-1 MELK Expression To Control Asymmetric Cell Division.Genetics. 2017 Aug;206(4):2069-2084. doi: 10.1534/genetics.117.202754. Epub 2017 Jun 26. Genetics. 2017. PMID: 28652378 Free PMC article.
-
Caenorhabditis elegans ced-3 Caspase Is Required for Asymmetric Divisions That Generate Cells Programmed To Die.Genetics. 2018 Nov;210(3):983-998. doi: 10.1534/genetics.118.301500. Epub 2018 Sep 7. Genetics. 2018. PMID: 30194072 Free PMC article.
-
Specification of neurons through asymmetric cell divisions.Curr Opin Neurobiol. 2010 Feb;20(1):44-9. doi: 10.1016/j.conb.2009.09.014. Epub 2009 Nov 4. Curr Opin Neurobiol. 2010. PMID: 19896361 Review.
-
The first cell cycle of the Caenorhabditis elegans embryo: spatial and temporal control of an asymmetric cell division.Results Probl Cell Differ. 2011;53:109-33. doi: 10.1007/978-3-642-19065-0_6. Results Probl Cell Differ. 2011. PMID: 21630143 Review.
Cited by
-
Neurogenesis in Caenorhabditis elegans.Genetics. 2024 Oct 7;228(2):iyae116. doi: 10.1093/genetics/iyae116. Genetics. 2024. PMID: 39167071 Free PMC article. Review.
References
-
- Afgan, E., Baker, D., Batut, B., van den Beek, M., Bouvier, D., Cech, M., Chilton, J., Clements, D., Coraor, N., Grüning, B. A.et al. (2018). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537-W544. 10.1093/nar/gky379 - DOI - PMC - PubMed
-
- Boveri, T. (1899). Die Entwickelung von Ascaris megalocephala mit besonderer Rücksicht auf die Kernverhältnisse. In Festschrift f. C. v. Kupffer, pp. 383-430. G. Fischer, Jena. 10.5962/bhl.title.9070 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

