Computational reproducibility of Jupyter notebooks from biomedical publications
- PMID: 38206590
- PMCID: PMC10783158
- DOI: 10.1093/gigascience/giad113
Computational reproducibility of Jupyter notebooks from biomedical publications
Abstract
Background: Jupyter notebooks facilitate the bundling of executable code with its documentation and output in one interactive environment, and they represent a popular mechanism to document and share computational workflows, including for research publications. The reproducibility of computational aspects of research is a key component of scientific reproducibility but has not yet been assessed at scale for Jupyter notebooks associated with biomedical publications.
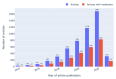
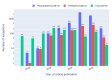
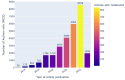
Approach: We address computational reproducibility at 2 levels: (i) using fully automated workflows, we analyzed the computational reproducibility of Jupyter notebooks associated with publications indexed in the biomedical literature repository PubMed Central. We identified such notebooks by mining the article's full text, trying to locate them on GitHub, and attempting to rerun them in an environment as close to the original as possible. We documented reproduction success and exceptions and explored relationships between notebook reproducibility and variables related to the notebooks or publications. (ii) This study represents a reproducibility attempt in and of itself, using essentially the same methodology twice on PubMed Central over the course of 2 years, during which the corpus of Jupyter notebooks from articles indexed in PubMed Central has grown in a highly dynamic fashion.
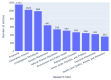
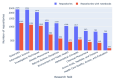
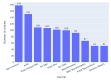
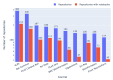
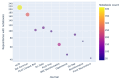
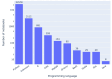
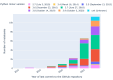
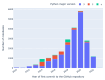
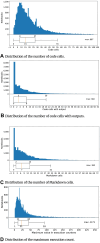
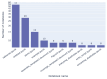
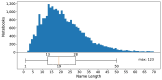
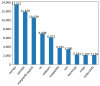
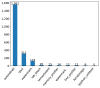
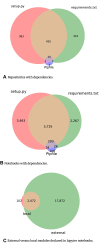
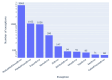
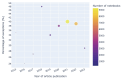
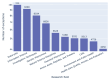
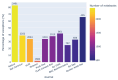
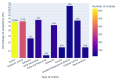
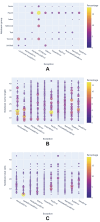
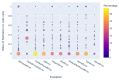
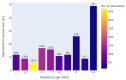
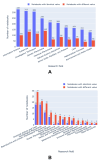
Results: Out of 27,271 Jupyter notebooks from 2,660 GitHub repositories associated with 3,467 publications, 22,578 notebooks were written in Python, including 15,817 that had their dependencies declared in standard requirement files and that we attempted to rerun automatically. For 10,388 of these, all declared dependencies could be installed successfully, and we reran them to assess reproducibility. Of these, 1,203 notebooks ran through without any errors, including 879 that produced results identical to those reported in the original notebook and 324 for which our results differed from the originally reported ones. Running the other notebooks resulted in exceptions.
Conclusions: We zoom in on common problems and practices, highlight trends, and discuss potential improvements to Jupyter-related workflows associated with biomedical publications.
Keywords: GitHub; Jupyter notebooks; PubMed Central; Python; computational reproducibility; dependency decay; workflow documentation.
© The Author(s) 2024. Published by Oxford University Press GigaScience.
Conflict of interest statement
The authors declare there are no competing interests.
Figures



Similar articles
-
Reproducible Bioconductor workflows using browser-based interactive notebooks and containers.J Am Med Inform Assoc. 2018 Jan 1;25(1):4-12. doi: 10.1093/jamia/ocx120. J Am Med Inform Assoc. 2018. PMID: 29092073 Free PMC article.
-
Understanding and improving the quality and reproducibility of Jupyter notebooks.Empir Softw Eng. 2021;26(4):65. doi: 10.1007/s10664-021-09961-9. Epub 2021 May 8. Empir Softw Eng. 2021. PMID: 33994841 Free PMC article.
-
A simple kit to use computational notebooks for more openness, reproducibility, and productivity in research.PLoS Comput Biol. 2022 Sep 15;18(9):e1010356. doi: 10.1371/journal.pcbi.1010356. eCollection 2022 Sep. PLoS Comput Biol. 2022. PMID: 36107931 Free PMC article.
-
Toward collaborative open data science in metabolomics using Jupyter Notebooks and cloud computing.Metabolomics. 2019 Sep 14;15(10):125. doi: 10.1007/s11306-019-1588-0. Metabolomics. 2019. PMID: 31522294 Free PMC article. Review.
-
Streamlining statistical reproducibility: NHLBI ORCHID clinical trial results reproduction.JAMIA Open. 2022 Jan 14;5(1):ooac001. doi: 10.1093/jamiaopen/ooac001. eCollection 2022 Apr. JAMIA Open. 2022. PMID: 35156003 Free PMC article. Review.
Cited by
-
Neuroimaging article reexecution and reproduction assessment system.Front Neuroinform. 2024 Jul 22;18:1376022. doi: 10.3389/fninf.2024.1376022. eCollection 2024. Front Neuroinform. 2024. PMID: 39104828 Free PMC article.
-
Distributed Collaboration for Data, Analysis Pipelines, and Results in Single-Cell Omics.bioRxiv [Preprint]. 2024 Jul 30:2024.07.30.605714. doi: 10.1101/2024.07.30.605714. bioRxiv. 2024. PMID: 39131282 Free PMC article. Preprint.
-
The five pillars of computational reproducibility: bioinformatics and beyond.Brief Bioinform. 2023 Sep 22;24(6):bbad375. doi: 10.1093/bib/bbad375. Brief Bioinform. 2023. PMID: 37870287 Free PMC article.
-
Best practices for data management and sharing in experimental biomedical research.Physiol Rev. 2024 Jul 1;104(3):1387-1408. doi: 10.1152/physrev.00043.2023. Epub 2024 Mar 7. Physiol Rev. 2024. PMID: 38451234 Free PMC article. Review.
-
Historical perspective and future directions: computational science in immuno-oncology.J Immunother Cancer. 2024 Jan 8;12(1):e008306. doi: 10.1136/jitc-2023-008306. J Immunother Cancer. 2024. PMID: 38191244 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

