Development and Validation of a Robust and Interpretable Early Triaging Support System for Patients Hospitalized With COVID-19: Predictive Algorithm Modeling and Interpretation Study
- PMID: 38206673
- PMCID: PMC10811577
- DOI: 10.2196/52134
Development and Validation of a Robust and Interpretable Early Triaging Support System for Patients Hospitalized With COVID-19: Predictive Algorithm Modeling and Interpretation Study
Abstract
Background: Robust and accurate prediction of severity for patients with COVID-19 is crucial for patient triaging decisions. Many proposed models were prone to either high bias risk or low-to-moderate discrimination. Some also suffered from a lack of clinical interpretability and were developed based on early pandemic period data. Hence, there has been a compelling need for advancements in prediction models for better clinical applicability.
Objective: The primary objective of this study was to develop and validate a machine learning-based Robust and Interpretable Early Triaging Support (RIETS) system that predicts severity progression (involving any of the following events: intensive care unit admission, in-hospital death, mechanical ventilation required, or extracorporeal membrane oxygenation required) within 15 days upon hospitalization based on routinely available clinical and laboratory biomarkers.
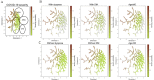
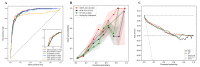
Methods: We included data from 5945 hospitalized patients with COVID-19 from 19 hospitals in South Korea collected between January 2020 and August 2022. For model development and external validation, the whole data set was partitioned into 2 independent cohorts by stratified random cluster sampling according to hospital type (general and tertiary care) and geographical location (metropolitan and nonmetropolitan). Machine learning models were trained and internally validated through a cross-validation technique on the development cohort. They were externally validated using a bootstrapped sampling technique on the external validation cohort. The best-performing model was selected primarily based on the area under the receiver operating characteristic curve (AUROC), and its robustness was evaluated using bias risk assessment. For model interpretability, we used Shapley and patient clustering methods.
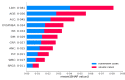
Results: Our final model, RIETS, was developed based on a deep neural network of 11 clinical and laboratory biomarkers that are readily available within the first day of hospitalization. The features predictive of severity included lactate dehydrogenase, age, absolute lymphocyte count, dyspnea, respiratory rate, diabetes mellitus, c-reactive protein, absolute neutrophil count, platelet count, white blood cell count, and saturation of peripheral oxygen. RIETS demonstrated excellent discrimination (AUROC=0.937; 95% CI 0.935-0.938) with high calibration (integrated calibration index=0.041), satisfied all the criteria of low bias risk in a risk assessment tool, and provided detailed interpretations of model parameters and patient clusters. In addition, RIETS showed potential for transportability across variant periods with its sustainable prediction on Omicron cases (AUROC=0.903, 95% CI 0.897-0.910).
Conclusions: RIETS was developed and validated to assist early triaging by promptly predicting the severity of hospitalized patients with COVID-19. Its high performance with low bias risk ensures considerably reliable prediction. The use of a nationwide multicenter cohort in the model development and validation implicates generalizability. The use of routinely collected features may enable wide adaptability. Interpretations of model parameters and patients can promote clinical applicability. Together, we anticipate that RIETS will facilitate the patient triaging workflow and efficient resource allocation when incorporated into a routine clinical practice.
Keywords: COVID-19; Omicron; SARS-CoV-2; SHAP; Shapley; biomarker; biomarkers; clustering; coronavirus; deep learning; early triaging; emergency; hospital admission; hospital admissions; hospitalization; hospitalizations; hospitalize; interpretability; machine learning; neural network; neural networks; predict; prediction; prediction model; predictive; prognosis; prognostic; prognostics; severity; triage; triaging.
©Sangwon Baek, Yeon joo Jeong, Yun-Hyeon Kim, Jin Young Kim, Jin Hwan Kim, Eun Young Kim, Jae-Kwang Lim, Jungok Kim, Zero Kim, Kyunga Kim, Myung Jin Chung. Originally published in the Journal of Medical Internet Research (https://www.jmir.org), 11.01.2024.
Conflict of interest statement
Conflicts of Interest: None declared.
Figures



Similar articles
-
The Development and Validation of Simplified Machine Learning Algorithms to Predict Prognosis of Hospitalized Patients With COVID-19: Multicenter, Retrospective Study.J Med Internet Res. 2022 Jan 21;24(1):e31549. doi: 10.2196/31549. J Med Internet Res. 2022. PMID: 34951865 Free PMC article.
-
Development and validation of an interpretable machine learning model for predicting in-hospital mortality for ischemic stroke patients in ICU.Int J Med Inform. 2025 Jun;198:105874. doi: 10.1016/j.ijmedinf.2025.105874. Epub 2025 Mar 9. Int J Med Inform. 2025. PMID: 40073651
-
Learning From Past Respiratory Infections to Predict COVID-19 Outcomes: Retrospective Study.J Med Internet Res. 2021 Feb 22;23(2):e23026. doi: 10.2196/23026. J Med Internet Res. 2021. PMID: 33534724 Free PMC article.
-
Prognostic models for newly-diagnosed chronic lymphocytic leukaemia in adults: a systematic review and meta-analysis.Cochrane Database Syst Rev. 2020 Jul 31;7(7):CD012022. doi: 10.1002/14651858.CD012022.pub2. Cochrane Database Syst Rev. 2020. PMID: 32735048 Free PMC article.
-
Developing and Testing Models for COVID-19 Health Outcomes [Internet].Washington (DC): Patient-Centered Outcomes Research Institute (PCORI); 2023 Mar. Washington (DC): Patient-Centered Outcomes Research Institute (PCORI); 2023 Mar. PMID: 37440675 Free Books & Documents. Review.
Cited by
-
Machine Learning for the Prediction of Acute Kidney Injury in Critically Ill Patients With Coronary Heart Disease: Algorithm Development and Validation.JMIR Med Inform. 2025 May 28;13:e72349. doi: 10.2196/72349. JMIR Med Inform. 2025. PMID: 40383933 Free PMC article.
-
Deep Learning-based Time-to-event Analysis of Depression and Asthma using the All of Us Research Program.AMIA Annu Symp Proc. 2025 May 22;2024:1186-1195. eCollection 2024. AMIA Annu Symp Proc. 2025. PMID: 40417537 Free PMC article.
-
Machine Learning and Artificial Intelligence for Infectious Disease Surveillance, Diagnosis, and Prognosis.Viruses. 2025 Jun 23;17(7):882. doi: 10.3390/v17070882. Viruses. 2025. PMID: 40733500 Free PMC article. Review.
-
Predicting COVID-19 severity in pediatric patients using machine learning: a comparative analysis of algorithms and ensemble methods.Sci Rep. 2025 Aug 8;15(1):29118. doi: 10.1038/s41598-025-15366-1. Sci Rep. 2025. PMID: 40781476 Free PMC article.
References
-
- WHO COVID-19 dashboard. World Health Organization. [2024-01-02]. https://data.who.int/dashboards/covid19/cases?n=c .
-
- Aleem A, Samad ABA, Vaqar S. StatPearls [Internet] Treasure Island, FL: StatPearls Publishing; 2023. Emerging variants of SARS-CoV-2 and novel therapeutics against coronavirus (COVID-19) - PubMed
-
- Lenharo M. WHO declares end to COVID-19's emergency phase. Nature. 2023. May 05, [2024-01-02]. https://www.nature.com/articles/d41586-023-01559-z . - PubMed
-
- Rahimi F, Darvishi M, Bezmin Abadi AT. 'The end' - or is it? Emergence of SARS-CoV-2 EG.5 and BA.2.86 subvariants. Future Virol. 2023 Sep;18(13):823. doi: 10.2217/fvl-2023-0150. https://europepmc.org/abstract/MED/37736262 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

