TRIM56 protects against nonalcoholic fatty liver disease by promoting the degradation of fatty acid synthase
- PMID: 38206764
- PMCID: PMC10904058
- DOI: 10.1172/JCI166149
TRIM56 protects against nonalcoholic fatty liver disease by promoting the degradation of fatty acid synthase
Abstract
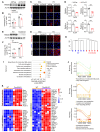
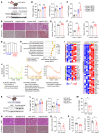
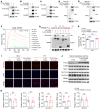
Nonalcoholic fatty liver disease (NAFLD) encompasses a disease continuum from simple steatosis to nonalcoholic steatohepatitis (NASH). However, there are currently no approved pharmacotherapies for NAFLD, although several drugs are in advanced stages of clinical development. Because of the complex pathophysiology and heterogeneity of NAFLD, the identification of potential therapeutic targets is clinically important. Here, we demonstrated that tripartite motif 56 (TRIM56) protein abundance was markedly downregulated in the livers of individuals with NAFLD and of mice fed a high-fat diet. Hepatocyte-specific ablation of TRIM56 exacerbated the progression of NAFLD, while hepatic TRIM56 overexpression suppressed it. Integrative analyses of interactome and transcriptome profiling revealed a pivotal role of TRIM56 in lipid metabolism and identified the lipogenesis factor fatty acid synthase (FASN) as a direct binding partner of TRIM56. TRIM56 directly interacted with FASN and triggered its K48-linked ubiquitination-dependent degradation. Finally, using artificial intelligence-based virtual screening, we discovered an orally bioavailable small-molecule inhibitor of FASN (named FASstatin) that potentiates TRIM56-mediated FASN ubiquitination. Therapeutic administration of FASstatin improved NAFLD and NASH pathologies in mice with an optimal safety, tolerability, and pharmacokinetics profile. Our findings provide proof of concept that targeting the TRIM56/FASN axis in hepatocytes may offer potential therapeutic avenues to treat NAFLD.
Keywords: Hepatology; Ubiquitin-proteosome system.
Figures






Similar articles
-
Fatty Acid Synthase-Suppressor Screening Identifies Sorting Nexin 8 as a Therapeutic Target for NAFLD.Hepatology. 2021 Nov;74(5):2508-2525. doi: 10.1002/hep.32045. Epub 2021 Sep 21. Hepatology. 2021. PMID: 34231239
-
Expression of fatty acid synthase in nonalcoholic fatty liver disease.Int J Clin Exp Pathol. 2010 Mar 25;3(5):505-14. Int J Clin Exp Pathol. 2010. PMID: 20606731 Free PMC article.
-
Mechanism of O-GlcNAcylation regulating liver lipid synthesis in mice through FASN.FASEB J. 2025 Feb 28;39(4):e70359. doi: 10.1096/fj.202402451RR. FASEB J. 2025. PMID: 39950311
-
A Comparison of the Gene Expression Profiles of Non-Alcoholic Fatty Liver Disease between Animal Models of a High-Fat Diet and Methionine-Choline-Deficient Diet.Molecules. 2022 Jan 27;27(3):858. doi: 10.3390/molecules27030858. Molecules. 2022. PMID: 35164140 Free PMC article. Review.
-
Recent advances in targeting the fatty acid biosynthetic pathway using fatty acid synthase inhibitors.Expert Opin Drug Discov. 2016 Dec;11(12):1187-1199. doi: 10.1080/17460441.2016.1245286. Epub 2016 Oct 18. Expert Opin Drug Discov. 2016. PMID: 27701891 Review.
Cited by
-
Ubiquitination and De-Ubiquitination in the Synthesis of Cow Milk Fat: Reality and Prospects.Molecules. 2024 Aug 29;29(17):4093. doi: 10.3390/molecules29174093. Molecules. 2024. PMID: 39274941 Free PMC article. Review.
-
Mechanism of Metabolic Dysfunction-associated Steatotic Liver Disease: Important role of lipid metabolism.J Clin Transl Hepatol. 2024 Sep 28;12(9):815-826. doi: 10.14218/JCTH.2024.00019. Epub 2024 Sep 3. J Clin Transl Hepatol. 2024. PMID: 39280069 Free PMC article. Review.
-
Comprehensive analysis of TRIM56's prognostic value and immune infiltration in Pan-Cancer.Sci Rep. 2025 Apr 21;15(1):13673. doi: 10.1038/s41598-025-97856-w. Sci Rep. 2025. PMID: 40258986 Free PMC article.
-
The clinical antiprotozoal drug halofuginone promotes weight loss by elevating GDF15 and FGF21.Sci Adv. 2025 Mar 28;11(13):eadt3142. doi: 10.1126/sciadv.adt3142. Epub 2025 Mar 26. Sci Adv. 2025. PMID: 40138418 Free PMC article.
-
Revisiting the role of GDF15 in atherosclerosis in mouse and human.Acta Pharmacol Sin. 2025 Apr 30. doi: 10.1038/s41401-025-01561-3. Online ahead of print. Acta Pharmacol Sin. 2025. PMID: 40307459
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

