Retinoic acid signaling regulates spatiotemporal specification of human green and red cones
- PMID: 38206904
- PMCID: PMC10783767
- DOI: 10.1371/journal.pbio.3002464
Retinoic acid signaling regulates spatiotemporal specification of human green and red cones
Abstract
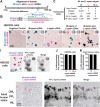
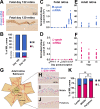
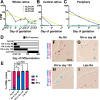
Trichromacy is unique to primates among placental mammals, enabled by blue (short/S), green (medium/M), and red (long/L) cones. In humans, great apes, and Old World monkeys, cones make a poorly understood choice between M and L cone subtype fates. To determine mechanisms specifying M and L cones, we developed an approach to visualize expression of the highly similar M- and L-opsin mRNAs. M-opsin was observed before L-opsin expression during early human eye development, suggesting that M cones are generated before L cones. In adult human tissue, the early-developing central retina contained a mix of M and L cones compared to the late-developing peripheral region, which contained a high proportion of L cones. Retinoic acid (RA)-synthesizing enzymes are highly expressed early in retinal development. High RA signaling early was sufficient to promote M cone fate and suppress L cone fate in retinal organoids. Across a human population sample, natural variation in the ratios of M and L cone subtypes was associated with a noncoding polymorphism in the NR2F2 gene, a mediator of RA signaling. Our data suggest that RA promotes M cone fate early in development to generate the pattern of M and L cones across the human retina.
Copyright: © 2024 Hadyniak et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
The role of opsin expression and apoptosis in determination of cone types in human retina.Exp Eye Res. 2004 Jun;78(6):1143-54. doi: 10.1016/j.exer.2004.01.004. Exp Eye Res. 2004. PMID: 15109921
-
Spatial and temporal expression of short, long/medium, or both opsins in human fetal cones.J Comp Neurol. 2000 Oct 2;425(4):545-59. J Comp Neurol. 2000. PMID: 10975879
-
Spatial and temporal expression of cone opsins during monkey retinal development.J Comp Neurol. 1997 Feb 3;378(1):117-34. J Comp Neurol. 1997. PMID: 9120051
-
Photopigment coexpression in mammals: comparative and developmental aspects.Histol Histopathol. 2005 Apr;20(2):551-74. doi: 10.14670/HH-20.551. Histol Histopathol. 2005. PMID: 15736061 Review.
-
Loss and gain of cone types in vertebrate ciliary photoreceptor evolution.Dev Biol. 2017 Nov 1;431(1):26-35. doi: 10.1016/j.ydbio.2017.08.038. Epub 2017 Sep 4. Dev Biol. 2017. PMID: 28882401 Review.
Cited by
-
Early retinoic acid signaling organizes the body axis and defines domains for the forelimb and eye.Curr Top Dev Biol. 2025;161:1-32. doi: 10.1016/bs.ctdb.2024.10.002. Epub 2024 Nov 8. Curr Top Dev Biol. 2025. PMID: 39870430 Free PMC article. Review.
-
Retinoid Synthesis Regulation by Retinal Cells in Health and Disease.Cells. 2024 May 18;13(10):871. doi: 10.3390/cells13100871. Cells. 2024. PMID: 38786093 Free PMC article.
-
Single cell dual-omic atlas of the human developing retina.Nat Commun. 2024 Aug 9;15(1):6792. doi: 10.1038/s41467-024-50853-5. Nat Commun. 2024. PMID: 39117640 Free PMC article.
-
Cell-cell interactions between transplanted retinal organoid cells and recipient tissues.Curr Opin Genet Dev. 2024 Dec;89:102277. doi: 10.1016/j.gde.2024.102277. Epub 2024 Nov 15. Curr Opin Genet Dev. 2024. PMID: 39549608 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

