The intricate cellular ecosystem of human peripheral veins as revealed by single-cell transcriptomic analysis
- PMID: 38206912
- PMCID: PMC10783777
- DOI: 10.1371/journal.pone.0296264
The intricate cellular ecosystem of human peripheral veins as revealed by single-cell transcriptomic analysis
Abstract
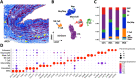
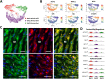
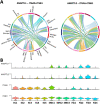
The venous system has been historically understudied despite its critical roles in blood distribution, heart function, and systemic immunity. This study dissects the microanatomy of upper arm veins at the single cell level, and how it relates to wall structure, remodeling processes, and inflammatory responses to injury. We applied single-cell RNA sequencing to 4 non-diseased human veins (3 basilic, 1 cephalic) obtained from organ donors, followed by bioinformatic and histological analyses. Unsupervised clustering of 20,006 cells revealed a complex ecosystem of endothelial cell (EC) types, smooth muscle cell (SMCs) and pericytes, various types of fibroblasts, and immune cell populations. The venous endothelium showed significant upregulation of cell adhesion genes, with arteriovenous zonation EC phenotypes highlighting the heterogeneity of vasa vasorum (VV) microvessels. Venous SMCs had atypical contractile phenotypes and showed widespread localization in the intima and media. MYH11+DESlo SMCs were transcriptionally associated with negative regulation of contraction and pro-inflammatory gene expression. MYH11+DEShi SMCs showed significant upregulation of extracellular matrix genes and pro-migratory mediators. Venous fibroblasts ranging from secretory to myofibroblastic phenotypes were 4X more abundant than SMCs and widely distributed throughout the wall. Fibroblast-derived angiopoietin-like factors were identified as versatile signaling hubs to regulate angiogenesis and SMC proliferation. An abundant monocyte/macrophage population was detected and confirmed by histology, including pro-inflammatory and homeostatic phenotypes, with cell counts positively correlated with age. Ligand-receptor interactome networks identified the venous endothelium in the main lumen and the VV as a niche for monocyte recruitment and infiltration. This study underscores the transcriptional uniqueness of venous cells and their relevance for vascular inflammation and remodeling processes. Findings from this study may be relevant for molecular investigations of upper arm veins used for vascular access creation, where single-cell analyses of cell composition and phenotypes are currently lacking.
Copyright: This is an open access article, free of all copyright, and may be freely reproduced, distributed, transmitted, modified, built upon, or otherwise used by anyone for any lawful purpose. The work is made available under the Creative Commons CC0 public domain dedication.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Single-Cell Analyses Offer Insights into the Different Remodeling Programs of Arteries and Veins.Cells. 2024 May 7;13(10):793. doi: 10.3390/cells13100793. Cells. 2024. PMID: 38786017 Free PMC article.
-
Novel Directly Reprogrammed Smooth Muscle Cells Promote Vascular Regeneration as Microvascular Mural Cells.Circulation. 2025 Apr 15;151(15):1076-1094. doi: 10.1161/CIRCULATIONAHA.124.070217. Epub 2025 Feb 13. Circulation. 2025. PMID: 39945059
-
Automated devices for identifying peripheral arterial disease in people with leg ulceration: an evidence synthesis and cost-effectiveness analysis.Health Technol Assess. 2024 Aug;28(37):1-158. doi: 10.3310/TWCG3912. Health Technol Assess. 2024. PMID: 39186036 Free PMC article.
-
Venous cutdown versus the Seldinger technique for placement of totally implantable venous access ports.Cochrane Database Syst Rev. 2016 Aug 21;2016(8):CD008942. doi: 10.1002/14651858.CD008942.pub2. Cochrane Database Syst Rev. 2016. PMID: 27544827 Free PMC article.
-
Single-Cell Transcriptomics and Lineage Tracing Unveil Parallels in Lymphatic Muscle and Venous Smooth Muscle Development, Identity, and Function.Arterioscler Thromb Vasc Biol. 2025 Jul;45(7):1207-1225. doi: 10.1161/ATVBAHA.125.322567. Epub 2025 May 15. Arterioscler Thromb Vasc Biol. 2025. PMID: 40371470 Free PMC article.
Cited by
-
Role of Perivascular Adipose Tissue in Vein Remodeling.Arterioscler Thromb Vasc Biol. 2025 May;45(5):576-584. doi: 10.1161/ATVBAHA.124.321692. Epub 2025 Mar 13. Arterioscler Thromb Vasc Biol. 2025. PMID: 40079141 Free PMC article. Review.
-
Single-Cell Analyses Offer Insights into the Different Remodeling Programs of Arteries and Veins.Cells. 2024 May 7;13(10):793. doi: 10.3390/cells13100793. Cells. 2024. PMID: 38786017 Free PMC article.
-
Characterization of Endothelial Cell Subclusters in Localized Scleroderma Skin with Single-Cell RNA Sequencing Identifies NOTCH Signaling Pathway.Int J Mol Sci. 2024 Sep 28;25(19):10473. doi: 10.3390/ijms251910473. Int J Mol Sci. 2024. PMID: 39408800 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

