Amyloid aggregates induced by the p53-R280T mutation lead to loss of p53 function in nasopharyngeal carcinoma
- PMID: 38212344
- PMCID: PMC10784298
- DOI: 10.1038/s41419-024-06429-8
Amyloid aggregates induced by the p53-R280T mutation lead to loss of p53 function in nasopharyngeal carcinoma
Abstract
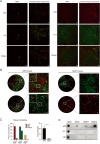
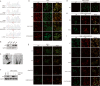
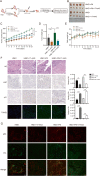
Nasopharyngeal carcinoma (NPC) is a malignant tumor that is highly prevalent in Southeast Asia, especially in South China. The pathogenesis of NPC is complex, and genetic alterations of tumor suppressors and proto-oncogenes play important roles in NPC carcinogenesis. p53 is unexpectedly highly expressed in NPC and possesses an uncommon mutation of R280T, which is different from a high frequency of hotspot mutations or low expression in other tumors. However, the mechanism of p53 loss of function and its correlation with R280T in NPC are still unclear. In this study, p53 amyloid aggregates were found to be widespread in NPC and can be mainly induced by the R280T mutation. Aggregated p53-R280T impeded its entry into the nucleus and was unable to initiate the transcription of downstream target genes, resulting in decreased NPC cell cycle arrest and apoptosis. In addition, NPC cells with p53-R280T amyloid aggregates also contributed aggressively to tumor growth in vivo. Transcriptome analysis suggested that p53 amyloid aggregation dysregulated major signaling pathways associated with the cell cycle, proliferation, apoptosis, and unfolded protein response (UPR). Further studies revealed that Hsp90, as a key molecular chaperone in p53 folding, was upregulated in NPC cells with p53-R280T aggregation, and the upregulated Hsp90 facilitated p53 aggregation in turn, forming positive feedback. Therefore, Hsp90 inhibitors could dissociate p53-R280T aggregation and restore the suppressor function of p53 in vitro and in vivo. In conclusion, our study demonstrated that p53-R280T may misfold to form aggregates with the help of Hsp90, resulting in the inability of sequestered p53 to initiate the transcription of downstream target genes. These results revealed a new mechanism for the loss of p53 function in NPC and provided novel mechanistic insight into NPC pathogenesis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Heterozygous p53-R280T Mutation Enhances the Oncogenicity of NPC Cells Through Activating PI3K-Akt Signaling Pathway.Front Oncol. 2020 Feb 5;10:104. doi: 10.3389/fonc.2020.00104. eCollection 2020. Front Oncol. 2020. PMID: 32117754 Free PMC article.
-
Mutant p53 confers chemoresistance by activating KMT5B-mediated DNA repair pathway in nasopharyngeal carcinoma.Cancer Lett. 2025 Aug 10;625:217736. doi: 10.1016/j.canlet.2025.217736. Epub 2025 Apr 30. Cancer Lett. 2025. PMID: 40316196
-
Cyclin B2 impairs the p53 signaling in nasopharyngeal carcinoma.BMC Cancer. 2024 Jan 2;24(1):25. doi: 10.1186/s12885-023-11768-4. BMC Cancer. 2024. PMID: 38166895 Free PMC article.
-
Functions and Roles of Long-Non-Coding RNAs in Human Nasopharyngeal Carcinoma.Cell Physiol Biochem. 2018;45(3):1191-1204. doi: 10.1159/000487451. Epub 2018 Feb 9. Cell Physiol Biochem. 2018. PMID: 29448252 Review.
-
Nutlin-3, A p53-Mdm2 Antagonist for Nasopharyngeal Carcinoma Treatment.Mini Rev Med Chem. 2018;18(2):173-183. doi: 10.2174/1389557517666170717125821. Mini Rev Med Chem. 2018. PMID: 28714398 Free PMC article. Review.
Cited by
-
Dominant-negative TP53 mutations potentiated by the HSF1-regulated proteostasis network.bioRxiv [Preprint]. 2024 Nov 3:2024.11.01.621414. doi: 10.1101/2024.11.01.621414. bioRxiv. 2024. PMID: 39554167 Free PMC article. Preprint.
-
Potential benefits of combined treatment with Hsp90 inhibitor AUY922 and cisplatin for overcoming drug resistance in nasopharyngeal carcinoma.Am J Cancer Res. 2025 Feb 15;15(2):533-545. doi: 10.62347/OSGO7209. eCollection 2025. Am J Cancer Res. 2025. PMID: 40084353 Free PMC article.
-
Ursolic acid induces apoptosis in nasopharyngeal carcinoma cells through the P53 signaling pathway: a network pharmacology and experimental validation study.Med Oncol. 2025 May 1;42(6):189. doi: 10.1007/s12032-025-02749-7. Med Oncol. 2025. PMID: 40310511
-
Δ133p53α and Δ160p53α isoforms of the tumor suppressor protein p53 exert dominant-negative effect primarily by co-aggregation.Elife. 2025 Jul 21;14:RP106469. doi: 10.7554/eLife.106469. Elife. 2025. PMID: 40690375 Free PMC article.
-
Strategies for Anticancer Treatment in p53-Mutated Head and Neck Squamous Cell Carcinoma.Biomedicines. 2025 May 10;13(5):1165. doi: 10.3390/biomedicines13051165. Biomedicines. 2025. PMID: 40426992 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- 81502542/National Natural Science Foundation of China (National Science Foundation of China)
- 81974074; 82172654/National Natural Science Foundation of China (National Science Foundation of China)
- 2023JJ40939/Hunan Provincial Science and Technology Department (Department of Science and Technology of Hunan Province)
- 2022JJ40797/Hunan Provincial Science and Technology Department (Department of Science and Technology of Hunan Province)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

