Genomic characterization of Pseudomonas spp. on food: implications for spoilage, antimicrobial resistance and human infection
- PMID: 38212698
- PMCID: PMC10782663
- DOI: 10.1186/s12866-023-03153-9
Genomic characterization of Pseudomonas spp. on food: implications for spoilage, antimicrobial resistance and human infection
Abstract
Background: Pseudomonas species are common on food, but their contribution to the antimicrobial resistance gene (ARG) burden within food or as a source of clinical infection is unknown. Pseudomonas aeruginosa is an opportunistic pathogen responsible for a wide range of infections and is often hard to treat due to intrinsic and acquired ARGs commonly carried by this species. This study aimed to understand the potential role of Pseudomonas on food as a reservoir of ARGs and to assess the presence of potentially clinically significant Pseudomonas aeruginosa strains on food. To achieve this, we assessed the genetic relatedness (using whole genome sequencing) and virulence of food-derived isolates to those collected from humans.
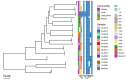
Results: A non-specific culturing approach for Pseudomonas recovered the bacterial genus from 28 of 32 (87.5%) retail food samples, although no P. aeruginosa was identified. The Pseudomonas species recovered were not clinically relevant, contained no ARGs and are likely associated with food spoilage. A specific culture method for P. aeruginosa resulted in the recovery of P. aeruginosa from 14 of 128 (11%) retail food samples; isolates contained between four and seven ARGs each and belonged to 16 sequence types (STs), four of which have been isolated from human infections. Food P. aeruginosa isolates from these STs demonstrated high similarity to human-derived isolates, differing by 41-312 single nucleotide polymorphisms (SNPs). There were diverse P. aeruginosa collected from the same food sample with distinct STs present on some samples and isolates belonging to the same ST differing by 19-67 SNPs. The Galleria mellonella infection model showed that 15 of 16 STs isolated from food displayed virulence between a low-virulence (PAO1) and a high virulence (PA14) control.
Conclusion: The most frequent Pseudomonas recovered from food examined in this study carried no ARGs and are more likely to play a role in food spoilage rather than infection. P. aeruginosa isolates likely to be able to cause human infections and with multidrug resistant genotypes are present on a relatively small but still substantial proportions of retail foods examined. Given the frequency of exposure, the potential contribution of food to the burden of P. aeruginosa infections in humans should be evaluated more closely.
Keywords: Antimicrobial resistance; Food; Pseudomonas aeruginosa; Pseudomonas spp.; Spoilage; Whole genome sequencing.
© 2024. The Author(s).
Conflict of interest statement
Competing interests. The authors declare no competing interests.
Figures



References
-
- Mellor GE, Bentley JA, Dykes GA. Evidence for a role of biosurfactants produced by Pseudomonas fluorescens in the spoilage of fresh aerobically stored chicken meat. Food Microbiol. 2011;28:1101–4. - PubMed
-
- Bloomfield SJ, Zomer AL, O’Grady J, Kay GL, Wain J, Janecko N, et al. Determination and quantification of microbial communities and antimicrobial resistance on food through host DNA-depleted metagenomics. Food Microbiol. 2023;110:1–12. - PubMed
-
- Walsh C, Duffy G, Nally P, O’Mahony R, McDowell DA, Fanning S. Transfer of ampicillin resistance from Salmonella Typhimurium DT104 to Escherichia coli K12 in food. Lett Appl Microbiol. 2008;46:210–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

