Exploring the relationship between metabolism and immune microenvironment in osteosarcoma based on metabolic pathways
- PMID: 38212768
- PMCID: PMC10785352
- DOI: 10.1186/s12929-024-00999-7
Exploring the relationship between metabolism and immune microenvironment in osteosarcoma based on metabolic pathways
Abstract
Background: Metabolic remodeling and changes in tumor immune microenvironment (TIME) in osteosarcoma are important factors affecting prognosis and treatment. However, the relationship between metabolism and TIME needs to be further explored.
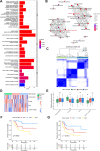
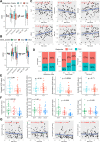
Methods: RNA-Seq data and clinical information of 84 patients with osteosarcoma from the TARGET database and an independent cohort from the GEO database were included in this study. The activity of seven metabolic super-pathways and immune infiltration levels were inferred in osteosarcoma patients. Metabolism-related genes (MRGs) were identified and different metabolic clusters and MRG-related gene clusters were identified using unsupervised clustering. Then the TIME differences between the different clusters were compared. In addition, an MRGs-based risk model was constructed and the role of a key risk gene, ST3GAL4, in osteosarcoma cells was explored using molecular biological experiments.
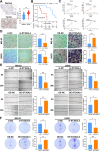
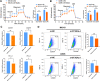
Results: This study revealed four key metabolic pathways in osteosarcoma, with vitamin and cofactor metabolism being the most relevant to prognosis and to TIME. Two metabolic pathway-related clusters (C1 and C2) were identified, with some differences in immune activating cell infiltration between the two clusters, and C2 was more likely to respond to two chemotherapeutic agents than C1. Three MRG-related gene clusters (GC1-3) were also identified, with significant differences in prognosis among the three clusters. GC2 and GC3 had higher immune cell infiltration than GC1. GC3 is most likely to respond to immune checkpoint blockade and to three commonly used clinical drugs. A metabolism-related risk model was developed and validated. The risk model has strong prognostic predictive power and the low-risk group has a higher level of immune infiltration than the high-risk group. Knockdown of ST3GAL4 significantly inhibited proliferation, migration, invasion and glycolysis of osteosarcoma cells and inhibited the M2 polarization of macrophages.
Conclusion: The metabolism of vitamins and cofactors is an important prognostic regulator of TIME in osteosarcoma, MRG-related gene clusters can well reflect changes in osteosarcoma TIME and predict chemotherapy and immunotherapy response. The metabolism-related risk model may serve as a useful prognostic predictor. ST3GAL4 plays a critical role in the progression, glycolysis, and TIME of osteosarcoma cells.
Keywords: Metabolism; Osteosarcoma; Prognosis; ST3GAL4; Treatment response; Tumor immune microenvironment; Vitamin and cofactor.
© 2024. The Author(s).
Conflict of interest statement
The authors declared no potential conflicts of interest.
Figures






Similar articles
-
A tumor microenvironment-based prognostic index for osteosarcoma.J Biomed Sci. 2023 Apr 13;30(1):23. doi: 10.1186/s12929-023-00917-3. J Biomed Sci. 2023. PMID: 37055822 Free PMC article.
-
Comprehensive analysis of prognostic tumor microenvironment-related genes in osteosarcoma patients.BMC Cancer. 2020 Aug 27;20(1):814. doi: 10.1186/s12885-020-07216-2. BMC Cancer. 2020. PMID: 32854645 Free PMC article.
-
Identification and characterization of aging/senescence-induced genes in osteosarcoma and predicting clinical prognosis.Front Immunol. 2022 Oct 5;13:997765. doi: 10.3389/fimmu.2022.997765. eCollection 2022. Front Immunol. 2022. PMID: 36275664 Free PMC article.
-
Signature constructed by glycolysis-immune-related genes can predict the prognosis of osteosarcoma patients.Invest New Drugs. 2022 Aug;40(4):818-830. doi: 10.1007/s10637-022-01228-4. Epub 2022 Apr 18. Invest New Drugs. 2022. PMID: 35435626 Review.
-
Targeting one-carbon metabolism for cancer immunotherapy.Clin Transl Med. 2024 Jan;14(1):e1521. doi: 10.1002/ctm2.1521. Clin Transl Med. 2024. PMID: 38279895 Free PMC article. Review.
Cited by
-
Multi-omic validation of the cuproptosis-sphingolipid metabolism network: modulating the immune landscape in osteosarcoma.Front Immunol. 2024 Jun 25;15:1424806. doi: 10.3389/fimmu.2024.1424806. eCollection 2024. Front Immunol. 2024. PMID: 38983852 Free PMC article.
-
Exploring the Role of Immune Cells in Glioma: Causal Associations and Clinical Implications.Int J Med Sci. 2025 Jun 12;22(12):2973-2991. doi: 10.7150/ijms.116560. eCollection 2025. Int J Med Sci. 2025. PMID: 40657381 Free PMC article.
-
Single-cell transcriptomic analysis of glioblastoma reveals pericytes contributing to the blood-brain-tumor barrier and tumor progression.MedComm (2020). 2024 Dec 4;5(12):e70014. doi: 10.1002/mco2.70014. eCollection 2024 Dec. MedComm (2020). 2024. PMID: 39640361 Free PMC article.
-
RFWD3 Reprograms Nucleotide Metabolism Through PHGDH to Induce Chemoresistance In Osteosarcoma.Adv Sci (Weinh). 2025 Apr;12(16):e2410937. doi: 10.1002/advs.202410937. Epub 2025 Feb 28. Adv Sci (Weinh). 2025. PMID: 40019400 Free PMC article.
-
Manipulating the cGAS-STING Axis: advancing innovative strategies for osteosarcoma therapeutics.Front Immunol. 2025 Feb 7;16:1539396. doi: 10.3389/fimmu.2025.1539396. eCollection 2025. Front Immunol. 2025. PMID: 39991153 Free PMC article. Review.
References
-
- Bielack SS, Kempf-Bielack B, Delling G, Exner GU, Flege S, Helmke K, et al. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J Clin Oncol. 2002;20:776–790. doi: 10.1200/jco.2002.20.3.776. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

