Genomic basis of seed colour in quinoa inferred from variant patterns using extreme gradient boosting
- PMID: 38213076
- PMCID: PMC11022794
- DOI: 10.1111/pbi.14267
Genomic basis of seed colour in quinoa inferred from variant patterns using extreme gradient boosting
Abstract
Quinoa is an agriculturally important crop species originally domesticated in the Andes of central South America. One of its most important phenotypic traits is seed colour. Seed colour variation is determined by contrasting abundance of betalains, a class of strong antioxidant and free radicals scavenging colour pigments only found in plants of the order Caryophyllales. However, the genetic basis for these pigments in seeds remains to be identified. Here we demonstrate the application of machine learning (extreme gradient boosting) to identify genetic variants predictive of seed colour. We show that extreme gradient boosting outperforms the classical genome-wide association approach. We provide re-sequencing and phenotypic data for 156 South American quinoa accessions and identify candidate genes potentially controlling betalain content in quinoa seeds. Genes identified include novel cytochrome P450 genes and known members of the betalain synthesis pathway, as well as genes annotated as being involved in seed development. Our work showcases the power of modern machine learning methods to extract biologically meaningful information from large sequencing data sets.
Keywords: betalain synthesis pathway; genome sequencing; genotype‐phenotype relationships; machine learning; quinoa; seed colour.
© 2023 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
None declared.
Figures

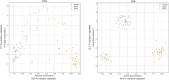


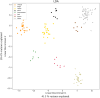
Similar articles
-
Identification, expression analysis of quinoa betalain biosynthesis genes and their role in seed germination and cold stress.Plant Signal Behav. 2023 Dec 31;18(1):2250891. doi: 10.1080/15592324.2023.2250891. Plant Signal Behav. 2023. PMID: 37616475 Free PMC article.
-
Root restriction accelerates genomic target identification in quinoa under controlled conditions.Physiol Plant. 2025 Mar-Apr;177(2):e70223. doi: 10.1111/ppl.70223. Physiol Plant. 2025. PMID: 40231839 Free PMC article.
-
Global investigation into the CqCYP76AD and CqDODA families in Chenopodium quinoa: Identification, evolutionary history, and their functional roles in betalain biosynthesis.Plant Physiol Biochem. 2025 Mar;220:109569. doi: 10.1016/j.plaphy.2025.109569. Epub 2025 Jan 27. Plant Physiol Biochem. 2025. PMID: 39892247
-
Progress on genomics and locus of important agronomic traits in Chenopodium quinoa.Yi Chuan. 2022 Nov 20;44(11):1009-1027. doi: 10.16288/j.yczz.22-289. Yi Chuan. 2022. PMID: 36384994 Review.
-
The evolution of betalain biosynthesis in Caryophyllales.New Phytol. 2019 Oct;224(1):71-85. doi: 10.1111/nph.15980. Epub 2019 Jul 19. New Phytol. 2019. PMID: 31172524 Review.
Cited by
-
Developing and validating a machine learning model to predict multidrug-resistant Klebsiella pneumoniae-related septic shock.Front Immunol. 2025 Jan 10;15:1539465. doi: 10.3389/fimmu.2024.1539465. eCollection 2024. Front Immunol. 2025. PMID: 39867898 Free PMC article.
-
Identification of CqCYP76AD5v1, a gene involved in betaxanthin biosynthesis in Chenopodium quinoa, and its product, betaxanthin, which inhibits amyloid-β aggregation.Plant Biotechnol (Tokyo). 2025 Jun 25;42(2):111-119. doi: 10.5511/plantbiotechnology.25.0122a. Plant Biotechnol (Tokyo). 2025. PMID: 40636428 Free PMC article.
-
From 'Farm to Fork': Exploring the Potential of Nutrient-Rich and Stress-Resilient Emergent Crops for Sustainable and Healthy Food in the Mediterranean Region in the Face of Climate Change Challenges.Plants (Basel). 2024 Jul 11;13(14):1914. doi: 10.3390/plants13141914. Plants (Basel). 2024. PMID: 39065441 Free PMC article. Review.
-
Genetic and environmental influences on fatty acid and tocopherol diversity in quinoa germplasm.Front Plant Sci. 2025 May 15;16:1541895. doi: 10.3389/fpls.2025.1541895. eCollection 2025. Front Plant Sci. 2025. PMID: 40453342 Free PMC article.
-
Variation analysis using random forests reveals domestication patterns and breeding trends in sugar beet.iScience. 2025 Jun 11;28(8):112835. doi: 10.1016/j.isci.2025.112835. eCollection 2025 Aug 15. iScience. 2025. PMID: 40740499 Free PMC article.
References
-
- van der Auwera, G. and O'Connor, B.D. (2020) Genomics in the Cloud: Using Docker, GATK, and WDL in Terra, 1st edn. Sebastopol, CA: O'Reilly Media.
-
- Bergstra, J. , Komer, B. , Eliasmith, C. , Yamins, D. and Cox, D.D. (2015) Hyperopt: a Python library for model selection and hyperparameter optimization. Comput. Sci. Discov. 8, 014008.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

