Recent advancements in targeted protein knockdown technologies-emerging paradigms for targeted therapy
- PMID: 38213543
- PMCID: PMC10776596
- DOI: 10.37349/etat.2023.00194
Recent advancements in targeted protein knockdown technologies-emerging paradigms for targeted therapy
Abstract
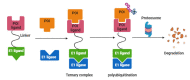
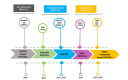
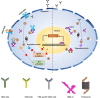
A generalized therapeutic strategy for various disease conditions, including cancer, is to deplete or inactivate harmful protein targets. Various forms of protein or gene silencing molecules, e.g., small molecule inhibitors, RNA interference (RNAi), and microRNAs (miRNAs) have been used against druggable targets. Over the past few years, targeted protein degradation (TPD) approaches have been developed for direct degradation of candidate proteins. Among the TPD approaches, proteolysis targeting chimeras (PROTACs) have emerged as one of the most promising approaches for the selective elimination of proteins via the ubiquitin-proteasome system. Other than PROTACs, TPD methods with potential therapeutic use include intrabody-mediated protein knockdown and tripartite motif-21 (TRIM-21) mediated TRIM-Away. In this review, protein knockdown approaches, their modes of action, and their advantages over conventional gene knockdown approaches are summarized. In cancers, disease-associated protein functions are often executed by specific post-translational modifications (PTMs). The role of TRIM-Away is highlighted in the direct knockdown of PTM forms of target proteins. Moreover, the application challenges and the prospective clinical use of TPD approaches in various diseases are also discussed.
Keywords: Protein knockdown; cancer; intrabodies; post-translational modification; proteolysis targeting chimeras; tripartite motif-21.
© The Author(s) 2023.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures
Similar articles
-
[Advances of targeted protein degradation technology and its applications in diseases therapy].Sheng Wu Gong Cheng Xue Bao. 2021 Nov 25;37(11):3915-3932. doi: 10.13345/j.cjb.200816. Sheng Wu Gong Cheng Xue Bao. 2021. PMID: 34841795 Review. Chinese.
-
Progress of small molecules for targeted protein degradation: PROTACs and other technologies.Drug Dev Res. 2023 Apr;84(2):337-394. doi: 10.1002/ddr.22026. Epub 2023 Jan 6. Drug Dev Res. 2023. PMID: 36606428 Review.
-
Targeted protein degradation in cancers: Orthodox PROTACs and beyond.Innovation (Camb). 2023 Mar 15;4(3):100413. doi: 10.1016/j.xinn.2023.100413. eCollection 2023 May 15. Innovation (Camb). 2023. PMID: 37033156 Free PMC article. Review.
-
The Potential of Proteolytic Chimeras as Pharmacological Tools and Therapeutic Agents.Molecules. 2020 Dec 16;25(24):5956. doi: 10.3390/molecules25245956. Molecules. 2020. PMID: 33339292 Free PMC article. Review.
-
Emerging Strategies in Proteolysis-Targeting Chimeras (PROTACs): Highlights from 2022.Int J Mol Sci. 2023 Mar 8;24(6):5190. doi: 10.3390/ijms24065190. Int J Mol Sci. 2023. PMID: 36982263 Free PMC article. Review.
Cited by
-
Post-Translational Modifications in Multiple Myeloma: Mechanisms of Drug Resistance and Therapeutic Opportunities.Biomolecules. 2025 May 12;15(5):702. doi: 10.3390/biom15050702. Biomolecules. 2025. PMID: 40427595 Free PMC article. Review.
-
The T-Box Transcription Factors TBX2 and TBX3 Are Molecular Targets of Piroctone Olamine in the Treatment of Pancreatic Cancer.J Cell Mol Med. 2025 Jul;29(14):e70736. doi: 10.1111/jcmm.70736. J Cell Mol Med. 2025. PMID: 40717225 Free PMC article.
-
Schottky barrier in pea-like Au@Bi2S3 nanoreactor enabling efficient photodynamic therapy of hepatocellular carcinoma.Mater Today Bio. 2025 Jun 19;33:102001. doi: 10.1016/j.mtbio.2025.102001. eCollection 2025 Aug. Mater Today Bio. 2025. PMID: 40642288 Free PMC article.
-
The treatment landscape of triple-negative breast cancer.Med Oncol. 2024 Aug 29;41(10):236. doi: 10.1007/s12032-024-02456-9. Med Oncol. 2024. PMID: 39210220 Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous
