Comparative evaluation of AlphaFold2 and disorder predictors for prediction of intrinsic disorder, disorder content and fully disordered proteins
- PMID: 38213902
- PMCID: PMC10782001
- DOI: 10.1016/j.csbj.2023.06.001
Comparative evaluation of AlphaFold2 and disorder predictors for prediction of intrinsic disorder, disorder content and fully disordered proteins
Abstract
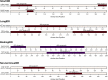
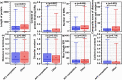
We expand studies of AlphaFold2 (AF2) in the context of intrinsic disorder prediction by comparing it against a broad selection of 20 accurate, popular and recently released disorder predictors. We use 25% larger benchmark dataset with 646 proteins and cover protein-level predictions of disorder content and fully disordered proteins. AF2-based disorder predictions secure a relatively high Area Under receiver operating characteristic Curve (AUC) of 0.77 and are statistically outperformed by several modern disorder predictors that secure AUCs around 0.8 with median runtime of about 20 s compared to 1200 s for AF2. Moreover, AF2 provides modestly accurate predictions of fully disordered proteins (F1 = 0.59 vs. 0.91 for the best disorder predictor) and disorder content (mean absolute error of 0.21 vs. 0.15). AF2 also generates statistically more accurate disorder predictions for about 20% of proteins that have relatively short sequences and a few disordered regions that tend to be located at the sequence termini, and which are absent of disordered protein-binding regions. Interestingly, AF2 and the most accurate disorder predictors rely on deep neural networks, suggesting that these models are useful for protein structure and disorder predictions.
Keywords: AlphaFold2; Deep learning; Disorder content; Fully disordered proteins; Intrinsic disorder; Intrinsically disordered protein; Prediction.
© 2023 The Authors.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Deep learning in prediction of intrinsic disorder in proteins.Comput Struct Biotechnol J. 2022 Mar 8;20:1286-1294. doi: 10.1016/j.csbj.2022.03.003. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35356546 Free PMC article. Review.
-
The Origin of Discrepancies between Predictions and Annotations in Intrinsically Disordered Proteins.Biomolecules. 2023 Sep 25;13(10):1442. doi: 10.3390/biom13101442. Biomolecules. 2023. PMID: 37892124 Free PMC article.
-
Comparative Assessment of Intrinsic Disorder Predictions with a Focus on Protein and Nucleic Acid-Binding Proteins.Biomolecules. 2020 Dec 4;10(12):1636. doi: 10.3390/biom10121636. Biomolecules. 2020. PMID: 33291838 Free PMC article. Review.
-
Accuracy of protein-level disorder predictions.Brief Bioinform. 2020 Sep 25;21(5):1509-1522. doi: 10.1093/bib/bbz100. Brief Bioinform. 2020. PMID: 31616935 Review.
-
How AlphaFold2 Predicts Conditionally Folding Regions Annotated in an Intrinsically Disordered Protein Database, IDEAL.Biology (Basel). 2023 Jan 25;12(2):182. doi: 10.3390/biology12020182. Biology (Basel). 2023. PMID: 36829461 Free PMC article.
Cited by
-
DescribePROT in 2023: more, higher-quality and experimental annotations and improved data download options.Nucleic Acids Res. 2024 Jan 5;52(D1):D426-D433. doi: 10.1093/nar/gkad985. Nucleic Acids Res. 2024. PMID: 37933852 Free PMC article.
-
Assessment of Disordered Linker Predictions in the CAID2 Experiment.Biomolecules. 2024 Feb 28;14(3):287. doi: 10.3390/biom14030287. Biomolecules. 2024. PMID: 38540707 Free PMC article.
-
Taxonomy-specific assessment of intrinsic disorder predictions at residue and region levels in higher eukaryotes, protists, archaea, bacteria and viruses.Comput Struct Biotechnol J. 2024 Apr 27;23:1968-1977. doi: 10.1016/j.csbj.2024.04.059. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 38765610 Free PMC article.
-
Characterization of alternative splicing during mammalian brain development reveals the extent of isoform diversity and potential effects on protein structural changes.Biol Open. 2024 Jul 15;13(10):bio061721. doi: 10.1242/bio.061721. Epub 2024 Oct 10. Biol Open. 2024. PMID: 39387301 Free PMC article.
-
Chaotic aging: intrinsically disordered proteins in aging-related processes.Cell Mol Life Sci. 2023 Aug 27;80(9):269. doi: 10.1007/s00018-023-04897-3. Cell Mol Life Sci. 2023. PMID: 37634152 Free PMC article.
References
-
- Oldfield C.J., et al. Introduction to intrinsically disordered proteins and regions. Intrinsically Disord Protein: Dyn, Bind, Funct. 2019:1–34.
-
- Habchi J., et al. Introducing protein intrinsic disorder. Chem Rev. 2014;114(13):6561–6588. - PubMed
-
- Ward J.J., et al. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337(3):635–645. - PubMed
-
- Xue B., Dunker A.K., Uversky V.N. Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J Biomol Struct Dyn. 2012;30(2):137–149. - PubMed
LinkOut - more resources
Full Text Sources
