Comparative evaluation of AlphaFold2 and disorder predictors for prediction of intrinsic disorder, disorder content and fully disordered proteins
- PMID: 38213902
- PMCID: PMC10782001
- DOI: 10.1016/j.csbj.2023.06.001
Comparative evaluation of AlphaFold2 and disorder predictors for prediction of intrinsic disorder, disorder content and fully disordered proteins
Abstract
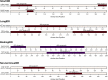
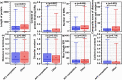
We expand studies of AlphaFold2 (AF2) in the context of intrinsic disorder prediction by comparing it against a broad selection of 20 accurate, popular and recently released disorder predictors. We use 25% larger benchmark dataset with 646 proteins and cover protein-level predictions of disorder content and fully disordered proteins. AF2-based disorder predictions secure a relatively high Area Under receiver operating characteristic Curve (AUC) of 0.77 and are statistically outperformed by several modern disorder predictors that secure AUCs around 0.8 with median runtime of about 20 s compared to 1200 s for AF2. Moreover, AF2 provides modestly accurate predictions of fully disordered proteins (F1 = 0.59 vs. 0.91 for the best disorder predictor) and disorder content (mean absolute error of 0.21 vs. 0.15). AF2 also generates statistically more accurate disorder predictions for about 20% of proteins that have relatively short sequences and a few disordered regions that tend to be located at the sequence termini, and which are absent of disordered protein-binding regions. Interestingly, AF2 and the most accurate disorder predictors rely on deep neural networks, suggesting that these models are useful for protein structure and disorder predictions.
Keywords: AlphaFold2; Deep learning; Disorder content; Fully disordered proteins; Intrinsic disorder; Intrinsically disordered protein; Prediction.
© 2023 The Authors.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Oldfield C.J., et al. Introduction to intrinsically disordered proteins and regions. Intrinsically Disord Protein: Dyn, Bind, Funct. 2019:1–34.
-
- Habchi J., et al. Introducing protein intrinsic disorder. Chem Rev. 2014;114(13):6561–6588. - PubMed
-
- Ward J.J., et al. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337(3):635–645. - PubMed
-
- Xue B., Dunker A.K., Uversky V.N. Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J Biomol Struct Dyn. 2012;30(2):137–149. - PubMed
LinkOut - more resources
Full Text Sources
