Atg8 family proteins, LIR/AIM motifs and other interaction modes
- PMID: 38214012
- PMCID: PMC7615515
- DOI: 10.1080/27694127.2023.2188523
Atg8 family proteins, LIR/AIM motifs and other interaction modes
Abstract
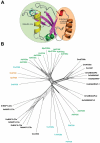
The Atg8 family of ubiquitin-like proteins play pivotal roles in autophagy and other processes involving vesicle fusion and transport where the lysosome/vacuole is the end station. Nuclear roles of Atg8 proteins are also emerging. Here, we review the structural and functional features of Atg8 family proteins and their protein-protein interaction modes in model organisms such as yeast, Arabidopsis, C. elegans and Drosophila to humans. Although varying in number of homologs, from one in yeast to seven in humans, and more than ten in some plants, there is a strong evolutionary conservation of structural features and interaction modes. The most prominent interaction mode is between the LC3 interacting region (LIR), also called Atg8 interacting motif (AIM), binding to the LIR docking site (LDS) in Atg8 homologs. There are variants of these motifs like "half-LIRs" and helical LIRs. We discuss details of the binding modes and how selectivity is achieved as well as the role of multivalent LIR-LDS interactions in selective autophagy. A number of LIR-LDS interactions are known to be regulated by phosphorylation. New methods to predict LIR motifs in proteins have emerged that will aid in discovery and analyses. There are also other interaction surfaces than the LDS becoming known where we presently lack detailed structural information, like the N-terminal arm region and the UIM-docking site (UDS). More interaction modes are likely to be discovered in future studies.
Keywords: AIM; Atg8; Autophagy; LDS; LIR; UDS; phosphorylation; protein-protein interaction.
Conflict of interest statement
Disclosure statement No potential conflict of interest was reported by the author(s).
Figures




References
-
- Matoba K, Kotani T, Tsutsumi A, Tsuji T, Mori T, Noshiro D, Sugita Y, Nomura N, Iwata S, Ohsumi Y, Fujimoto T, Nakatogawa H, Kikkawa M, Noda NN.. Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion. Nat Struct Mol Biol. 2020. Dec;27(12):1185–1193. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
