Prefusion stabilization of the Hendra and Langya virus F proteins
- PMID: 38214525
- PMCID: PMC10878279
- DOI: 10.1128/jvi.01372-23
Prefusion stabilization of the Hendra and Langya virus F proteins
Abstract
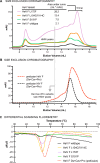
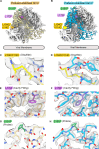
Nipah virus (NiV) and Hendra virus (HeV) are pathogenic paramyxoviruses that cause mild-to-severe disease in humans. As members of the Henipavirus genus, NiV and HeV use an attachment (G) glycoprotein and a class I fusion (F) glycoprotein to invade host cells. The F protein rearranges from a metastable prefusion form to an extended postfusion form to facilitate host cell entry. Prefusion NiV F elicits higher neutralizing antibody titers than postfusion NiV F, indicating that stabilization of prefusion F may aid vaccine development. A combination of amino acid substitutions (L104C/I114C, L172F, and S191P) is known to stabilize NiV F in its prefusion conformation, although the extent to which substitutions transfer to other henipavirus F proteins is not known. Here, we perform biophysical and structural studies to investigate the mechanism of prefusion stabilization in F proteins from three henipaviruses: NiV, HeV, and Langya virus (LayV). Three known stabilizing substitutions from NiV F transfer to HeV F and exert similar structural and functional effects. One engineered disulfide bond, located near the fusion peptide, is sufficient to stabilize the prefusion conformations of both HeV F and LayV F. Although LayV F shares low overall sequence identity with NiV F and HeV F, the region around the fusion peptide exhibits high sequence conservation across all henipaviruses. Our findings indicate that substitutions targeting this site of conformational change might be applicable to prefusion stabilization of other henipavirus F proteins and support the use of NiV as a prototypical pathogen for henipavirus vaccine antigen design.IMPORTANCEPathogenic henipaviruses such as Nipah virus (NiV) and Hendra virus (HeV) cause respiratory symptoms, with severe cases resulting in encephalitis, seizures, and coma. The work described here shows that the NiV and HeV fusion (F) proteins share common structural features with the F protein from an emerging henipavirus, Langya virus (LayV). Sequence alignment alone was sufficient to predict which known prefusion-stabilizing amino acid substitutions from NiV F would stabilize the prefusion conformations of HeV F and LayV F. This work also reveals an unexpected oligomeric interface shared by prefusion HeV F and NiV F. Together, these advances lay a foundation for future antigen design targeting henipavirus F proteins. In this way, Nipah virus can serve as a prototypical pathogen for the development of protective vaccines and monoclonal antibodies to prepare for potential henipavirus outbreaks.
Keywords: Hendra; Langya; Nipah; electron microscopy; paramyxovirus; prefusion; structure-based vaccine design.
Conflict of interest statement
J.S.M., R.J.L., and B.S.G. are inventors on U.S. patent application no. 62/714,230 ("Nipah virus Immunogens and Their Use"). B.S.G. and R.J.L. are inventors on US patent application no. 63/315,934 (filed 2 March 2022, "Monoclonal Antibodies to Nipah Virus F Protein and Their Use").
Figures



References
-
- Chua KB, Bellini WJ, Rota PA, Harcourt BH, Tamin A, Lam SK, Ksiazek TG, Rollin PE, Zaki SR, Shieh W, Goldsmith CS, Gubler DJ, Roehrig JT, Eaton B, Gould AR, Olson J, Field H, Daniels P, Ling AE, Peters CJ, Anderson LJ, Mahy BW. 2000. Nipah virus: a recently emergent deadly paramyxovirus. Science 288:1432–1435. doi:10.1126/science.288.5470.1432 - DOI - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Research Materials

