Opportunities and limitations of genomics for diagnosing bedaquiline-resistant tuberculosis: a systematic review and individual isolate meta-analysis
- PMID: 38215766
- PMCID: PMC11072239
- DOI: 10.1016/S2666-5247(23)00317-8
Opportunities and limitations of genomics for diagnosing bedaquiline-resistant tuberculosis: a systematic review and individual isolate meta-analysis
Abstract
Background: Clinical bedaquiline resistance predominantly involves mutations in mmpR5 (Rv0678). However, mmpR5 resistance-associated variants (RAVs) have a variable relationship with phenotypic Mycobacterium tuberculosis resistance. We did a systematic review to assess the maximal sensitivity of sequencing bedaquiline resistance-associated genes and evaluate the association between RAVs and phenotypic resistance, using traditional and machine-based learning techniques.
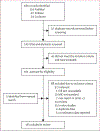
Methods: We screened public databases for articles published from database inception until Oct 31, 2022. Eligible studies performed sequencing of at least mmpR5 and atpE on clinically sourced M tuberculosis isolates and measured bedaquiline minimum inhibitory concentrations (MICs). A bias risk scoring tool was used to identify bias. Individual genetic mutations and corresponding MICs were aggregated, and odds ratios calculated to determine association of mutations with resistance. Machine-based learning methods were used to define test characteristics of parsimonious sets of diagnostic RAVs, and mmpR5 mutations were mapped to the protein structure to highlight mechanisms of resistance. This study was registered in the PROSPERO database (CRD42022346547).
Findings: 18 eligible studies were identified, comprising 975 M tuberculosis isolates containing at least one potential RAV (mutation in mmpR5, atpE, atpB, or pepQ), with 201 (20·6%) showing phenotypic bedaquiline resistance. 84 (29·5%) of 285 resistant isolates had no candidate gene mutation. Sensitivity and positive predictive value of taking an any mutation approach was 69% and 14%, respectively. 13 mutations, all in mmpR5, had a significant association with a resistant MIC (adjusted p<0·05). Gradient-boosted machine classifier models for predicting intermediate or resistant and resistant phenotypes both had receiver operator characteristic c statistic of 0·73 (95% CI 0·70-0·76). Frameshift mutations clustered in the α1 helix DNA-binding domain, and substitutions in the α2 and α3 helix hinge region and in the α4 helix-binding domain.
Interpretation: Sequencing candidate genes is insufficiently sensitive to diagnose clinical bedaquiline resistance, but where identified, some mutations should be assumed to be associated with resistance. Genomic tools are most likely to be effective in combination with rapid phenotypic diagnostics. This study was limited by selective sampling in contributing studies and only considering single genetic loci as causative of resistance.
Funding: Francis Crick Institute and National Institute of Allergy and Infectious Diseases at the National Institutes of Health.
Copyright © 2023 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of interests CN declares a grant from the Academy of Medical Sciences, consultancy fees from Columbia University, and travel award from the British Thoracic Society. MJC, MHL, and MO declare grants from the National Institutes of Health/National Institute of Allergy and Infectious Diseases (NIH/NIAID). All other authors declare no competing interests.
Figures


Update of
-
Opportunities and limitations of genomics for diagnosing bedaquiline-resistant tuberculosis: an individual isolate metaanalysis.medRxiv [Preprint]. 2023 May 5:2023.05.04.23289023. doi: 10.1101/2023.05.04.23289023. medRxiv. 2023. Update in: Lancet Microbe. 2024 Feb;5(2):e164-e172. doi: 10.1016/S2666-5247(23)00317-8. PMID: 37205550 Free PMC article. Updated. Preprint.
References
-
- WHO. Consolidated guidelines on tuberculosis. Module 4: treatment—drug-resistant tuberculosis treatment, 2022 update. World Health Organization. 2022. https://www.who.int/publications/i/item/9789240063129 (accessed Nov 15, 2023). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

