The E3 ubiquitin ligase Itch regulates death receptor and cholesterol trafficking to affect TRAIL-mediated apoptosis
- PMID: 38216558
- PMCID: PMC10786908
- DOI: 10.1038/s41419-023-06417-4
The E3 ubiquitin ligase Itch regulates death receptor and cholesterol trafficking to affect TRAIL-mediated apoptosis
Abstract
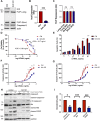
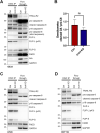
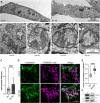
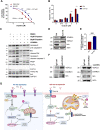
The activation of apoptosis signalling by TRAIL (TNF-related apoptosis-inducing ligand) through receptor binding is a fundamental mechanism of cell death induction and is often perturbed in cancer cells to enhance their cell survival and treatment resistance. Ubiquitination plays an important role in the regulation of TRAIL-mediated apoptosis, and here we investigate the role of the E3 ubiquitin ligase Itch in TRAIL-mediated apoptosis in oesophageal cancer cells. Knockdown of Itch expression results in resistance to TRAIL-induced apoptosis, caspase-8 activation, Bid cleavage and also promotes cisplatin resistance. Whilst the assembly of the death-inducing signalling complex (DISC) at the plasma membrane is not perturbed relative to the control, TRAIL-R2 is mis-localised in the Itch-knockdown cells. Further, we observe significant changes to mitochondrial morphology alongside an increased cholesterol content. Mitochondrial cholesterol is recognised as an important anti-apoptotic agent in cancer. Cells treated with a drug that increases mitochondrial cholesterol levels, U18666A, shows a protection from TRAIL-induced apoptosis, reduced caspase-8 activation, Bid cleavage and cisplatin resistance. We demonstrate that Itch knockdown cells are less sensitive to a Bcl-2 inhibitor, show impaired activation of Bax, cytochrome c release and an enhanced stability of the cholesterol transfer protein STARD1. We identify a novel protein complex composed of Itch, the mitochondrial protein VDAC2 and STARD1. We propose a mechanism where Itch regulates the stability of STARD1. An increase in STARD1 expression enhances cholesterol import to mitochondria, which inhibits Bax activation and cytochrome c release. Many cancer types display high mitochondrial cholesterol levels, and oesophageal adenocarcinoma tumours show a correlation between chemotherapy resistance and STARD1 expression which is supported by our findings. This establishes an important role for Itch in regulation of extrinsic and intrinsic apoptosis, mitochondrial cholesterol levels and provides insight to mechanisms that contribute to TRAIL, Bcl-2 inhibitor and cisplatin resistance in cancer cells.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Clorennec CL, Lazrek Y, Dubreuil O, Sampaio C, Larbouret C, Lanotte R, et al. ITCH-dependent proteasomal degradation of c-FLIP induced by the anti-HER3 antibody 9F7-F11 promotes DR5/caspase 8-mediated apoptosis of tumor cells. Cell Commun Signal. 2019;17:106. doi: 10.1186/s12964-019-0413-8. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

