Global emergence of a hypervirulent carbapenem-resistant Escherichia coli ST410 clone
- PMID: 38216585
- PMCID: PMC10786849
- DOI: 10.1038/s41467-023-43854-3
Global emergence of a hypervirulent carbapenem-resistant Escherichia coli ST410 clone
Abstract
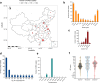
Carbapenem-resistant Escherichia coli (CREC) ST410 has recently emerged as a major global health problem. Here, we report a shift in CREC prevalence in Chinese hospitals between 2017 and 2021 with ST410 becoming the most commonly isolated sequence type. Genomic analysis identifies a hypervirulent CREC ST410 clone, B5/H24RxC, which caused two separate outbreaks in a children's hospital. It may have emerged from the previously characterised B4/H24RxC in 2006 and has been isolated in ten other countries from 2015 to 2021. Compared with B4/H24RxC, B5/H24RxC lacks the blaOXA-181-bearing X3 plasmid, but carries a F-type plasmid containing blaNDM-5. Most of B5/H24RxC also carry a high pathogenicity island and a novel O-antigen gene cluster. We find that B5/H24RxC grew faster in vitro and is more virulent in vivo. The identification of this newly emerged but already globally disseminated hypervirulent CREC clone, highlights the ongoing evolution of ST410 towards increased resistance and virulence.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

