The origins of the Guinness stout yeast
- PMID: 38216745
- PMCID: PMC10786833
- DOI: 10.1038/s42003-023-05587-3
The origins of the Guinness stout yeast
Abstract
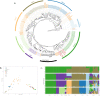
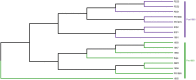
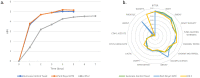
Beer is made via the fermentation of an aqueous extract predominantly composed of malted barley flavoured with hops. The transforming microorganism is typically a single strain of Saccharomyces cerevisiae, and for the majority of major beer brands the yeast strain is a unique component. The present yeast used to make Guinness stout brewed in Dublin, Ireland, can be traced back to 1903, but its origins are unknown. To that end, we used Illumina and Nanopore sequencing to generate whole-genome sequencing data for a total of 22 S. cerevisiae yeast strains: 16 from the Guinness collection and 6 other historical Irish brewing. The origins of the Guinness yeast were determined with a SNP-based analysis, demonstrating that the Guinness strains occupy a distinct group separate from other historical Irish brewing yeasts. Assessment of chromosome number, copy number variation and phenotypic evaluation of key brewing attributes established Guinness yeast-specific SNPs but no specific chromosomal amplifications. Our analysis also demonstrated the effects of yeast storage on phylogeny. Altogether, our results suggest that the Guinness yeast used today is related to the first deposited Guinness yeast; the 1903 Watling Laboratory Guinness yeast.
© 2024. The Author(s).
Conflict of interest statement
Daniel Kerrruish, Jessica Kearns, Eibhlin Colgan, and Sandra Stelma are employees of Diageo Ireland, the owners of Guinness. Chris Boulton is employed as a consultant by Diageo Ireland. All other authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

