Robustness evaluations of pathway activity inference methods on gene expression data
- PMID: 38216898
- PMCID: PMC10785356
- DOI: 10.1186/s12859-024-05632-w
Robustness evaluations of pathway activity inference methods on gene expression data
Abstract
Background: With the exponential growth of high-throughput technologies, multiple pathway analysis methods have been proposed to estimate pathway activities from gene expression profiles. These pathway activity inference methods can be divided into two main categories: non-Topology-Based (non-TB) and Pathway Topology-Based (PTB) methods. Although some review and survey articles discussed the topic from different aspects, there is a lack of systematic assessment and comparisons on the robustness of these approaches.
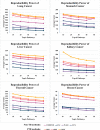
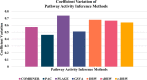
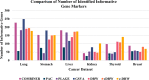
Results: Thus, this study presents comprehensive robustness evaluations of seven widely used pathway activity inference methods using six cancer datasets based on two assessments. The first assessment seeks to investigate the robustness of pathway activity in pathway activity inference methods, while the second assessment aims to assess the robustness of risk-active pathways and genes predicted by these methods. The mean reproducibility power and total number of identified informative pathways and genes were evaluated. Based on the first assessment, the mean reproducibility power of pathway activity inference methods generally decreased as the number of pathway selections increased. Entropy-based Directed Random Walk (e-DRW) distinctly outperformed other methods in exhibiting the greatest reproducibility power across all cancer datasets. On the other hand, the second assessment shows that no methods provide satisfactory results across datasets.
Conclusion: However, PTB methods generally appear to perform better in producing greater reproducibility power and identifying potential cancer markers compared to non-TB methods.
Keywords: Cancer classification; Literature validation; Pathway activity inference; Pathway analysis; PubMed text data mining; Reproducibility power; Robustness.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
An Entropy-Based Directed Random Walk for Cancer Classification Using Gene Expression Data Based on Bi-Random Walk on Two Separated Networks.Genes (Basel). 2023 Feb 24;14(3):574. doi: 10.3390/genes14030574. Genes (Basel). 2023. PMID: 36980844 Free PMC article.
-
Topologically inferring risk-active pathways toward precise cancer classification by directed random walk.Bioinformatics. 2013 Sep 1;29(17):2169-77. doi: 10.1093/bioinformatics/btt373. Epub 2013 Jul 10. Bioinformatics. 2013. PMID: 23842813
-
MICRAT: a novel algorithm for inferring gene regulatory networks using time series gene expression data.BMC Syst Biol. 2018 Dec 14;12(Suppl 7):115. doi: 10.1186/s12918-018-0635-1. BMC Syst Biol. 2018. PMID: 30547796 Free PMC article.
-
Identifying significantly impacted pathways: a comprehensive review and assessment.Genome Biol. 2019 Oct 9;20(1):203. doi: 10.1186/s13059-019-1790-4. Genome Biol. 2019. PMID: 31597578 Free PMC article. Review.
-
Evaluating network inference methods in terms of their ability to preserve the topology and complexity of genetic networks.Semin Cell Dev Biol. 2016 Mar;51:44-52. doi: 10.1016/j.semcdb.2016.01.012. Epub 2016 Feb 3. Semin Cell Dev Biol. 2016. PMID: 26851626 Review.
Cited by
-
Competing endogenous RNAs (ceRNAs) and drug resistance to cancer therapy.Cancer Drug Resist. 2024 Sep 25;7:37. doi: 10.20517/cdr.2024.66. eCollection 2024. Cancer Drug Resist. 2024. PMID: 39403602 Free PMC article. Review.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

