DNI-MDCAP: improvement of causal MiRNA-disease association prediction based on deep network imputation
- PMID: 38216907
- PMCID: PMC10785389
- DOI: 10.1186/s12859-024-05644-6
DNI-MDCAP: improvement of causal MiRNA-disease association prediction based on deep network imputation
Abstract
Background: MiRNAs are involved in the occurrence and development of many diseases. Extensive literature studies have demonstrated that miRNA-disease associations are stratified and encompass ~ 20% causal associations. Computational models that predict causal miRNA-disease associations provide effective guidance in identifying novel interpretations of disease mechanisms and potential therapeutic targets. Although several predictive models for miRNA-disease associations exist, it is still challenging to discriminate causal miRNA-disease associations from non-causal ones. Hence, there is a pressing need to develop an efficient prediction model for causal miRNA-disease association prediction.
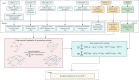
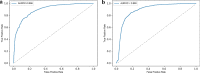
Results: We developed DNI-MDCAP, an improved computational model that incorporated additional miRNA similarity metrics, deep graph embedding learning-based network imputation and semi-supervised learning framework. Through extensive predictive performance evaluation, including tenfold cross-validation and independent test, DNI-MDCAP showed excellent performance in identifying causal miRNA-disease associations, achieving an area under the receiver operating characteristic curve (AUROC) of 0.896 and 0.889, respectively. Regarding the challenge of discriminating causal miRNA-disease associations from non-causal ones, DNI-MDCAP exhibited superior predictive performance compared to existing models MDCAP and LE-MDCAP, reaching an AUROC of 0.870. Wilcoxon test also indicated significantly higher prediction scores for causal associations than for non-causal ones. Finally, the potential causal miRNA-disease associations predicted by DNI-MDCAP, exemplified by diabetic nephropathies and hsa-miR-193a, have been validated by recently published literature, further supporting the reliability of the prediction model.
Conclusions: DNI-MDCAP is a dedicated tool to specifically distinguish causal miRNA-disease associations with substantially improved accuracy. DNI-MDCAP is freely accessible at http://www.rnanut.net/DNIMDCAP/ .
Keywords: Causal miRNA-disease association prediction; Deep graph embedding; Network imputation; miRNA; miRNA-disease association.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
LE-MDCAP: A Computational Model to Prioritize Causal miRNA-Disease Associations.Int J Mol Sci. 2021 Dec 19;22(24):13607. doi: 10.3390/ijms222413607. Int J Mol Sci. 2021. PMID: 34948403 Free PMC article.
-
A Computational Model to Predict the Causal miRNAs for Diseases.Front Genet. 2019 Oct 3;10:935. doi: 10.3389/fgene.2019.00935. eCollection 2019. Front Genet. 2019. PMID: 31632446 Free PMC article. Review.
-
EOESGC: predicting miRNA-disease associations based on embedding of embedding and simplified graph convolutional network.BMC Med Inform Decis Mak. 2021 Nov 16;21(1):319. doi: 10.1186/s12911-021-01671-y. BMC Med Inform Decis Mak. 2021. PMID: 34789236 Free PMC article.
-
Identification of miRNA-disease associations via deep forest ensemble learning based on autoencoder.Brief Bioinform. 2022 May 13;23(3):bbac104. doi: 10.1093/bib/bbac104. Brief Bioinform. 2022. PMID: 35325038
-
A Survey of Deep Learning for Detecting miRNA- Disease Associations: Databases, Computational Methods, Challenges, and Future Directions.IEEE/ACM Trans Comput Biol Bioinform. 2024 May-Jun;21(3):328-347. doi: 10.1109/TCBB.2024.3351752. Epub 2024 Jun 5. IEEE/ACM Trans Comput Biol Bioinform. 2024. PMID: 38194377 Review.
References
-
- Guo FH, Guan YN, Guo JJ, Zhang LJ, Qiu JJ, Ji Y, Chen AF, Jing Q. Single-Cell transcriptome analysis reveals embryonic endothelial heterogeneity at spatiotemporal level and multifunctions of microRNA-126 in mice. Arterioscler Thromb Vasc Biol. 2022;42(3):326–342. doi: 10.1161/ATVBAHA.121.317093. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

