XRCC1: a potential prognostic and immunological biomarker in LGG based on systematic pan-cancer analysis
- PMID: 38217545
- PMCID: PMC10817400
- DOI: 10.18632/aging.205426
XRCC1: a potential prognostic and immunological biomarker in LGG based on systematic pan-cancer analysis
Abstract
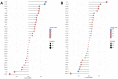
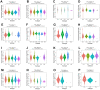
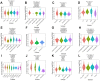
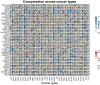
X-ray repair cross-complementation group 1 (XRCC1) is a pivotal contributor to base excision repair, and its dysregulation has been implicated in the oncogenicity of various human malignancies. However, a comprehensive pan-cancer analysis investigating the prognostic value, immunological functions, and epigenetic associations of XRCC1 remains lacking. To address this knowledge gap, we conducted a systematic investigation employing bioinformatics techniques across 33 cancer types. Our analysis encompassed XRCC1 expression levels, prognostic and diagnostic implications, epigenetic profiles, immune and molecular subtypes, Tumor Mutation Burden (TMB), Microsatellite Instability (MSI), immune checkpoints, and immune infiltration, leveraging data from TCGA, GTEx, CELL, Human Protein Atlas, Ualcan, and cBioPortal databases. Notably, XRCC1 displayed both positive and negative correlations with prognosis across different tumors. Epigenetic analysis revealed associations between XRCC1 expression and DNA methylation patterns in 10 cancer types, as well as enhanced phosphorylation. Furthermore, XRCC1 expression demonstrated associations with TMB and MSI in the majority of tumors. Interestingly, XRCC1 gene expression exhibited a negative correlation with immune cell infiltration levels, except for a positive correlation with M1 and M2 macrophages and monocytes in most cancers. Additionally, we observed significant correlations between XRCC1 and immune checkpoint gene expression levels. Lastly, our findings implicated XRCC1 in DNA replication and repair processes, shedding light on the precise mechanisms underlying its oncogenic effects. Overall, our study highlights the potential of XRCC1 as a prognostic and immunological pan-cancer biomarker, thereby offering a novel target for tumor immunotherapy.
Keywords: X-ray repair cross-complementation group 1; immune infiltration; pan-cancer; prognosis; tumor microenvironment.
Conflict of interest statement
Figures






Similar articles
-
Systematic Pan-Cancer Analysis Identifies TREM2 as an Immunological and Prognostic Biomarker.Front Immunol. 2021 Feb 17;12:646523. doi: 10.3389/fimmu.2021.646523. eCollection 2021. Front Immunol. 2021. PMID: 33679809 Free PMC article.
-
Multiomic characterization, immunological and prognostic potential of SMAD3 in pan-cancer and validation in LIHC.Sci Rep. 2025 Jan 3;15(1):657. doi: 10.1038/s41598-024-84553-3. Sci Rep. 2025. PMID: 39753728 Free PMC article.
-
NOX4 as an emerging prognostic and immunological biomarker across pan-cancer analyses.Sci Rep. 2025 May 29;15(1):18812. doi: 10.1038/s41598-025-03499-2. Sci Rep. 2025. PMID: 40442199 Free PMC article.
-
RREB1 could act as an immunological and prognostic biomarker: From comprehensive analysis to osteosarcoma validation.Int Immunopharmacol. 2024 Dec 25;143(Pt 1):113312. doi: 10.1016/j.intimp.2024.113312. Epub 2024 Oct 14. Int Immunopharmacol. 2024. PMID: 39405927
-
Golgi scaffold protein PAQR11 in pan-cancer landscape: A comprehensive bioinformatics exploration of expression patterns, prognostic significance, and potential immunological function.Heliyon. 2025 Jan 4;11(2):e41724. doi: 10.1016/j.heliyon.2025.e41724. eCollection 2025 Jan 30. Heliyon. 2025. PMID: 39906812 Free PMC article. Review.
Cited by
-
Integrative single-cell and spatial transcriptomics uncover ELK4-mediated mechanisms in NDUFAB1+ tumor cells driving gastric cancer progression, metabolic reprogramming, and immune evasion.Front Immunol. 2025 Jul 4;16:1591123. doi: 10.3389/fimmu.2025.1591123. eCollection 2025. Front Immunol. 2025. PMID: 40688093 Free PMC article.
-
Exploring the role of Disulfidptosis in glioma progression: insights into tumor heterogeneity and therapeutic potential through single-cell RNA sequencing.Discov Oncol. 2024 Dec 23;15(1):829. doi: 10.1007/s12672-024-01685-8. Discov Oncol. 2024. PMID: 39714742 Free PMC article.
References
-
- Dodgshun AJ, Fukuoka K, Edwards M, Bianchi VJ, Das A, Sexton-Oates A, Larouche V, Vanan MI, Lindhorst S, Yalon M, Mason G, Crooks B, Constantini S, et al.. Germline-driven replication repair-deficient high-grade gliomas exhibit unique hypomethylation patterns. Acta Neuropathol. 2020; 140:765–76. 10.1007/s00401-020-02209-8 - DOI - PubMed
-
- Jayaraj R, Raymond G, Krishnan S, Tzou KS, Baxi S, Ram MR, Govind SK, Chandramoorthy HC, Abu-Khzam FN, Shaw P. Clinical Theragnostic Potential of Diverse miRNA Expressions in Prostate Cancer: A Systematic Review and Meta-Analysis. Cancers (Basel). 2020; 12:1199. 10.3390/cancers12051199 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

