Natural variation in the zinc-finger-encoding exon of Prdm9 affects hybrid sterility phenotypes in mice
- PMID: 38217871
- PMCID: PMC10917509
- DOI: 10.1093/genetics/iyae004
Natural variation in the zinc-finger-encoding exon of Prdm9 affects hybrid sterility phenotypes in mice
Abstract
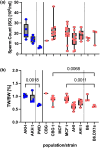
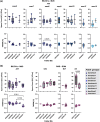
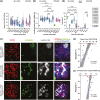
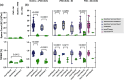
PRDM9-mediated reproductive isolation was first described in the progeny of Mus musculus musculus (MUS) PWD/Ph and Mus musculus domesticus (DOM) C57BL/6J inbred strains. These male F1 hybrids fail to complete chromosome synapsis and arrest meiosis at prophase I, due to incompatibilities between the Prdm9 gene and hybrid sterility locus Hstx2. We identified 14 alleles of Prdm9 in exon 12, encoding the DNA-binding domain of the PRDM9 protein in outcrossed wild mouse populations from Europe, Asia, and the Middle East, 8 of which are novel. The same allele was found in all mice bearing introgressed t-haplotypes encompassing Prdm9. We asked whether 7 novel Prdm9 alleles in MUS populations and the t-haplotype allele in 1 MUS and 3 DOM populations induce Prdm9-mediated reproductive isolation. The results show that only combinations of the dom2 allele of DOM origin and the MUS msc1 allele ensure complete infertility of intersubspecific hybrids in outcrossed wild populations and inbred mouse strains examined so far. The results further indicate that MUS mice may share the erasure of PRDM9msc1 binding motifs in populations with different Prdm9 alleles, which implies that erased PRDM9 binding motifs may be uncoupled from their corresponding Prdm9 alleles at the population level. Our data corroborate the model of Prdm9-mediated hybrid sterility beyond inbred strains of mice and suggest that sterility alleles of Prdm9 may be rare.
Keywords: Hstx2; Mus musculus; Prdm9; t-haplotype; asynapsis; fertility; reproductive isolation.
© The Author(s) 2024. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest The author(s) declare no conflicts of interest.
Figures



References
-
- Altemose N, Noor N, Bitoun E, Tumian A, Imbeault M, Chapman JR, Aricescu AR, Myers SR. 2017b. Human PRDM9 can bind and activate promoters, and other zinc-finger proteins associate with reduced recombination in cis. bioRxiv. 10.1101/144295, preprint: not peer reviewed. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

