Gene knockout of RNA binding motif 5 in the brain alters RIMS2 protein homeostasis in the cerebellum and Hippocampus and exacerbates behavioral deficits after a TBI in mice
- PMID: 38218585
- PMCID: PMC11178365
- DOI: 10.1016/j.expneurol.2024.114690
Gene knockout of RNA binding motif 5 in the brain alters RIMS2 protein homeostasis in the cerebellum and Hippocampus and exacerbates behavioral deficits after a TBI in mice
Abstract
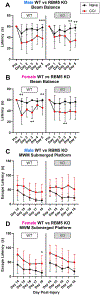
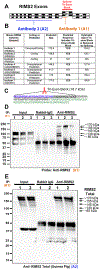
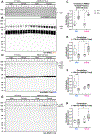
RNA binding motif 5 (RBM5) is a tumor suppressor in cancer but its role in the brain is unclear. We used conditional gene knockout (KO) mice to test if RBM5 inhibition in the brain affects chronic cortical brain tissue survival or function after a controlled cortical impact (CCI) traumatic brain injury (TBI). RBM5 KO decreased baseline contralateral hemispheric volume (p < 0.0001) and exacerbated ipsilateral tissue loss at 21 d after CCI in male mice vs. wild type (WT) (p = 0.0019). CCI injury, but not RBM5 KO, impaired beam balance performance (0-5d post-injury) and swim speed on the Morris Water Maze (MWM) (19-20d) (p < 0.0001). RBM5 KO was associated with mild learning impairment in female mice (p = 0.0426), reflected as a modest increase in escape latency early in training (14-18d post-injury). However, KO did not affect spatial memory at 19d post-injury in male or in female mice but it was impaired by CCI in females (p = 0.0061). RBM5 KO was associated with impaired visual function in male mice on the visible platform test at 20d post-injury (p = 0.0256). To explore signaling disturbances in KOs related to behavior, we first cross-referenced known brain-specific RBM5-regulated gene targets with genes in the curated RetNet database that impact vision. We then performed a secondary literature search on RBM5-regulated genes with a putative role in hippocampal function. Regulating synaptic membrane exocytosis 2 (RIMS) 2 was identified as a gene of interest because it regulates both vision and hippocampal function. Immunoprecipitation and western blot confirmed protein expression of a novel ~170 kDa RIMS2 variant in the cerebellum, and in the hippocampus, it was significantly increased in KO vs WT (p < 0.0001), and in a sex-dependent manner (p = 0.0390). Furthermore, male KOs had decreased total canonical RIMS2 levels in the cerebellum (p = 0.0027) and hippocampus (p < 0.0001), whereas female KOs had increased total RIMS1 levels in the cerebellum (p = 0.0389). In summary, RBM5 modulates brain function in mammals. Future work is needed to test if RBM5 dependent regulation of RIMS2 splicing effects vision and cognition, and to verify potential sex differences on behavior in a larger cohort of mice.
Keywords: Blindness; Cerebellum; Hippocampus; Learning; RBM5; RIMS2; Splicing; TBI; Vision.
Copyright © 2023. Published by Elsevier Inc.
Conflict of interest statement
Declaration of competing interest TCJ and PMK are inventors on a USPTO patent (No. 9610266).
Figures





References
-
- Elkin LA, Kay M, Higgins JJ, Wobbrock JO, 2021. An Aligned Rank Transform Procedure for Multifactor Contrast Tests, The 34th Annual ACM Symposium on User Interface Software and Technology. Association for Computing Machinery, Virtual Event, USA, pp. 754–768.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

