USP43 stabilizes c-Myc to promote glycolysis and metastasis in bladder cancer
- PMID: 38218970
- PMCID: PMC10787741
- DOI: 10.1038/s41419-024-06446-7
USP43 stabilizes c-Myc to promote glycolysis and metastasis in bladder cancer
Abstract
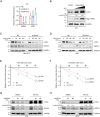
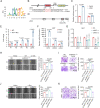
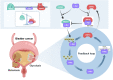
A hallmark of tumor cells, including bladder cancer (BLCA) cells, is metabolic reprogramming toward aerobic glycolysis (Warburg effect). The classical oncogene MYC, which is crucial in regulating glycolysis, is amplified and activated in BLCA. However, direct targeting of the c-Myc oncoprotein, which regulates glycolytic metabolism, presents great challenges and necessitates the discovery of a more clarified regulatory mechanism to develop selective targeted therapy. In this study, a siRNA library targeting deubiquitinases identified a candidate enzyme named USP43, which may regulate glycolytic metabolism and c-Myc transcriptional activity. Further investigation using functional assays and molecular studies revealed a USP43/c-Myc positive feedback loop that contributes to the progression of BLCA. Moreover, USP43 stabilizes c-Myc by deubiquitinating c-Myc at K148 and K289 primarily through deubiquitinase activity. Additionally, upregulation of USP43 protein in BLCA increased the chance of interaction with c-Myc and interfered with FBXW7 access and degradation of c-Myc. These findings suggest that USP43 is a potential therapeutic target for indirectly targeting glycolytic metabolism and the c-Myc oncoprotein consequently enhancing the efficacy of bladder cancer treatment.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

