Multiple Immune and Genetic Mechanisms Contribute to Cmv5s-Driven Susceptibility and Tissue Damage during Acute Murine Cytomegalovirus Infection
- PMID: 38224204
- PMCID: PMC10922835
- DOI: 10.4049/jimmunol.2300648
Multiple Immune and Genetic Mechanisms Contribute to Cmv5s-Driven Susceptibility and Tissue Damage during Acute Murine Cytomegalovirus Infection
Abstract
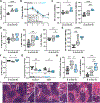
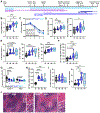
The MHC class I molecule H-2Dk conveys resistance to acute murine CMV infection in both C57L (H-2Dk transgenic) and MA/My mice. M.H2k/b mice are on an MA/My background aside from a C57L-derived region spanning the MHC (Cmv5s), which diminishes this resistance and causes significant spleen histopathology. To hone in on the effector elements within the Cmv5s interval, we generated several Cmv5-recombinant congenic mouse strains and screened them in vivo, allowing us to narrow the phenotype-associated interval >6-fold and segment the genetic mechanism to at least two independent loci within the MHC region. In addition, we sought to further characterize the Cmv5s-associated phenotypes in their temporal appearance and potential direct relationship to viral load. To this end, we found that Cmv5s histopathology and NK cell activation could not be fully mirrored in the MA/My mice with increased viral dose, and that marginal zone destruction was the first apparent Cmv5s phenotype, being reliably quantified as early as 2 d postinfection in the M.H2k/b mice, prior to divergence in viral load, weight loss, or NK cell phenotype. Finally, we further dissect NK cell involvement, finding no intrinsic differences in NK cell function, despite increased upregulation of activation markers and checkpoint receptors. In conclusion, these data dissect the genetic and immunologic underpinnings of Cmv5 and reveal a model in which polymorphism within the MHC region of the genome leads to the development of tissue damage and corrupts protective NK cell immunity during acute viral infection.
Copyright © 2024 by The American Association of Immunologists, Inc.
Figures





Similar articles
-
Inflammation and Macrophage Loss Mark Increased Susceptibility in a Genetic Model of Acute Viral Infection-Induced Tissue Damage.J Immunol. 2024 Sep 15;213(6):853-864. doi: 10.4049/jimmunol.2400116. J Immunol. 2024. PMID: 39046317
-
Genomic Modifiers of Natural Killer Cells, Immune Responsiveness and Lymphoid Tissue Remodeling Together Increase Host Resistance to Viral Infection.PLoS Pathog. 2016 Feb 4;12(2):e1005419. doi: 10.1371/journal.ppat.1005419. eCollection 2016 Feb. PLoS Pathog. 2016. PMID: 26845690 Free PMC article.
-
Requisite H2k role in NK cell-mediated resistance in acute murine cytomegalovirus-infected MA/My mice.J Immunol. 2005 Nov 15;175(10):6820-8. doi: 10.4049/jimmunol.175.10.6820. J Immunol. 2005. PMID: 16272339
-
Natural killer cell licensing during viral infection.Adv Exp Med Biol. 2011;780:37-44. doi: 10.1007/978-1-4419-5632-3_4. Adv Exp Med Biol. 2011. PMID: 21842363 Review.
-
NK cell recognition of mouse cytomegalovirus-infected cells.Curr Top Microbiol Immunol. 2006;298:183-206. doi: 10.1007/3-540-27743-9_10. Curr Top Microbiol Immunol. 2006. PMID: 16329187 Review.
Cited by
-
Inflammation and Macrophage Loss Mark Increased Susceptibility in a Genetic Model of Acute Viral Infection-Induced Tissue Damage.J Immunol. 2024 Sep 15;213(6):853-864. doi: 10.4049/jimmunol.2400116. J Immunol. 2024. PMID: 39046317
References
-
- Lanier LL 2005. NK Cell Recognition. Annu Rev Immunol. 23:225–274. - PubMed
-
- Kim S, Poursine-Laurent J, Truscott SM, Lybarger L, Song Y, Yang L, French AR, Sunwoo JB, Lemieux S, Hansen TH, Yokoyama WM. 2005. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 436:709–713. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

