dCas9/CRISPR-based methylation of O-6-methylguanine-DNA methyltransferase enhances chemosensitivity to temozolomide in malignant glioma
- PMID: 38224404
- PMCID: PMC10824881
- DOI: 10.1007/s11060-023-04531-z
dCas9/CRISPR-based methylation of O-6-methylguanine-DNA methyltransferase enhances chemosensitivity to temozolomide in malignant glioma
Abstract
Background: Malignant glioma carries a poor prognosis despite current therapeutic modalities. Standard of care therapy consists of surgical resection, fractionated radiotherapy concurrently administered with temozolomide (TMZ), a DNA-alkylating chemotherapeutic agent, followed by adjuvant TMZ. O-6-methylguanine-DNA methyltransferase (MGMT), a DNA repair enzyme, removes alkylated lesions from tumor DNA, thereby promoting chemoresistance. MGMT promoter methylation status predicts responsiveness to TMZ; patients harboring unmethylated MGMT (~60% of glioblastoma) have a poorer prognosis with limited treatment benefits from TMZ.
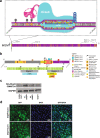
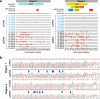
Methods: Via lentiviral-mediated delivery into LN18 glioma cells, we employed deactivated Cas9-CRISPR technology to target the MGMT promoter and enhancer regions for methylation, as mediated by the catalytic domain of the methylation enzyme DNMT3A. Methylation patterns were examined at a clonal level in regions containing Differentially Methylation Regions (DMR1, DMR2) and the Methylation Specific PCR (MSP) region used for clinical assessment of MGMT methylation status. Correlative studies of genomic and transcriptomic effects of dCas9/CRISPR-based methylation were performed via Illumina 850K methylation array platform and bulk RNA-Seq analysis.
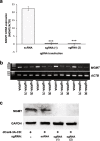
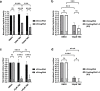
Results: We used the dCas9/DNMT3A catalytic domain to achieve targeted MGMT methylation at specific CpG clusters in the vicinity of promoter, enhancer, DMRs and MSP regions. Consequently, we observed MGMT downregulation and enhanced glioma chemosensitivity in survival assays in vitro, with minimal off-target effects.
Conclusion: dCas9/CRISPR is a viable method of epigenetic editing, using the DNMT3A catalytic domain. This study provides initial proof-of-principle for CRISPR technology applications in malignant glioma, laying groundwork for subsequent translational studies, with implications for future epigenetic editing-based clinical applications.
Keywords: CRISPR therapeutics; Chemoresistance; Epigenetics; MGMT; Malignant glioma/glioblastoma (GBM); Methylation.
© 2024. The Author(s).
Conflict of interest statement
SZR, TL, STP, and AL have a U.S. Provisional Patent Application No. 63/429348 entitled TARGETED EPIGENETIC EDITING AS NOVEL THERAPY FOR MALIGNANT GLIOMA, filed on 12/1/2022. Official patent filing is currently in process, as of the time of manuscript submission. TJP, BSCE and BMK declare no potential conflicts of interest. LML is a board member and stock holder of ClearPoint Neuro, on the medical advisory board of InSightec, a paid consultant for ImmPact Bio, and has received research funding through UCLA from Northwest Biotherapeutics. PLN has contracts with UCLA for the Brain Tumor Program with BMS, Chimerix, Erasca, Springsworks, and Recursion. TFC is cofounder, major stock holder, consultant and board member of Katmai Pharmaceuticals, member of the board and paid consultant for the 501c3 Global Coalition for Adaptive Research, holds stock in Chimerix and receives milestone payments and possible future royalties, member of the scientific advisory board for Break Through Cancer, member of the scientific advisory board for Cure Brain Cancer Foundation, has provided paid consulting services to Sagimet, Clinical Care Options, Ideology Health, Servier, Jubilant, Immvira, Gan & Lee, BrainStorm, Katmai, Sapience, Inovio, Vigeo Therapeutics, DNATrix, Tyme, SDP, Novartis, Roche, Kintara, Bayer, Merck, Boehinger Ingelheim, VBL, Amgen, Kiyatec, Odonate Therapeutics QED, Medefield, Pascal Biosciences, Bayer, Tocagen, Karyopharm, GW Pharma, Abbvie, VBI, Deciphera, VBL, Agios, Genocea, Celgene, Puma, Lilly, BMS, Cortice, Wellcome Trust, Novocure, Novogen, Boston Biomedical, Sunovion, Human Longevity, Insys, ProNai, Pfizer, Notable labs, Medqia Trizel, Medscape and has contracts with UCLA for the Brain Tumor Program with Oncovir, Merck, Oncoceutics, Novartis, Amgen, Abbvie, DNAtrix, Beigene, BMS, AstraZeneca, Kazia, Agios, Boston Biomedical, Deciphera, Tocagen, Orbus, AstraZeneca, Karyopharm. The Regents of the University of California (TFC employer) has licensed intellectual property co-invented by TFC to Katmai Pharmaceuticals.
Figures

Similar articles
-
Enhancing Temozolomide (TMZ) chemosensitivity using CRISPR-dCas9-mediated downregulation of O6-methylguanine DNA methyltransferase (MGMT).J Neurooncol. 2024 Aug;169(1):129-135. doi: 10.1007/s11060-024-04708-0. Epub 2024 May 19. J Neurooncol. 2024. PMID: 38762829
-
Epigenetic Activation of TUSC3 Sensitizes Glioblastoma to Temozolomide Independent of MGMT Promoter Methylation Status.Int J Mol Sci. 2023 Oct 14;24(20):15179. doi: 10.3390/ijms242015179. Int J Mol Sci. 2023. PMID: 37894860 Free PMC article.
-
Effect of lomeguatrib-temozolomide combination on MGMT promoter methylation and expression in primary glioblastoma tumor cells.Tumour Biol. 2013 Jun;34(3):1935-47. doi: 10.1007/s13277-013-0738-7. Epub 2013 Mar 22. Tumour Biol. 2013. PMID: 23519841
-
Anti-glioma therapy with temozolomide and status of the DNA-repair gene MGMT.Anticancer Res. 2009 Nov;29(11):4845-54. Anticancer Res. 2009. PMID: 20032445 Review.
-
Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity.J Clin Oncol. 2008 Sep 1;26(25):4189-99. doi: 10.1200/JCO.2007.11.5964. J Clin Oncol. 2008. PMID: 18757334 Review.
Cited by
-
Two decades of progress in glioma methylation research: the rise of temozolomide resistance and immunotherapy insights.Front Neurosci. 2024 Sep 2;18:1440756. doi: 10.3389/fnins.2024.1440756. eCollection 2024. Front Neurosci. 2024. PMID: 39286478 Free PMC article.
-
Cell-Based Glioma Models for Anticancer Drug Screening: From Conventional Adherent Cell Cultures to Tumor-Specific Three-Dimensional Constructs.Cells. 2024 Dec 17;13(24):2085. doi: 10.3390/cells13242085. Cells. 2024. PMID: 39768176 Free PMC article. Review.
-
Epigenetic Alterations in Glioblastoma Multiforme as Novel Therapeutic Targets: A Scoping Review.Int J Mol Sci. 2025 Jun 12;26(12):5634. doi: 10.3390/ijms26125634. Int J Mol Sci. 2025. PMID: 40565099 Free PMC article.
-
Evolution of Molecular Biomarkers and Precision Molecular Therapeutic Strategies in Glioblastoma.Cancers (Basel). 2024 Oct 29;16(21):3635. doi: 10.3390/cancers16213635. Cancers (Basel). 2024. PMID: 39518074 Free PMC article. Review.
-
Multiplexed epigenetic memory editing using CRISPRoff sensitizes glioblastoma to chemotherapy.Neuro Oncol. 2025 Jul 30;27(6):1443-1457. doi: 10.1093/neuonc/noaf055. Neuro Oncol. 2025. PMID: 39998382 Free PMC article.

