Identification of key genes and functional enrichment analysis of liver fibrosis in nonalcoholic fatty liver disease through weighted gene co-expression network analysis
- PMID: 38224712
- PMCID: PMC10788356
- DOI: 10.5808/gi.23051
Identification of key genes and functional enrichment analysis of liver fibrosis in nonalcoholic fatty liver disease through weighted gene co-expression network analysis
Abstract
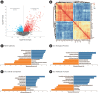
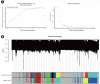
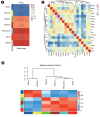
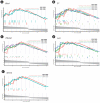
Nonalcoholic fatty liver disease (NAFLD) is a common type of chronic liver disease, with severity levels ranging from nonalcoholic fatty liver to nonalcoholic steatohepatitis (NASH). The extent of liver fibrosis indicates the severity of NASH and the risk of liver cancer. However, the mechanism underlying NASH development, which is important for early screening and intervention, remains unclear. Weighted gene co-expression network analysis (WGCNA) is a useful method for identifying hub genes and screening specific targets for diseases. In this study, we utilized an mRNA dataset of the liver tissues of patients with NASH and conducted WGCNA for various stages of liver fibrosis. Subsequently, we employed two additional mRNA datasets for validation purposes. Gene set enrichment analysis (GSEA) was conducted to analyze gene function enrichment. Through WGCNA and subsequent analyses, complemented by validation using two additional datasets, we identified five genes (BICC1, C7, EFEMP1, LUM, and STMN2) as hub genes. GSEA analysis indicated that gene sets associated with liver metabolism and cholesterol homeostasis were uniformly downregulated. BICC1, C7, EFEMP1, LUM, and STMN2 were identified as hub genes of NASH, and were all related to liver metabolism, NAFLD, NASH, and related diseases. These hub genes might serve as potential targets for the early screening and treatment of NASH.
Keywords: functional enrichment analysis; gene set enrichment analysis; hub genes; liver fibrosis; nonalcoholic fatty liver disease; weighted gene co-expression network analysis.
Conflict of interest statement
No potential conflict of interest relevant to this article was reported.
Figures


References
-
- Ahmed A, Wong RJ, Harrison SA. Nonalcoholic fatty liver disease review: diagnosis, treatment, and outcomes. Clin Gastroenterol Hepatol. 2015;13:2062–2070. - PubMed
-
- Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2018;15:11–20. - PubMed
-
- Marengo A, Rosso C, Bugianesi E. Liver cancer: connections with obesity, fatty liver, and cirrhosis. Annu Rev Med. 2016;67:103–117. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
