Large scale plasma proteomics identifies novel proteins and protein networks associated with heart failure development
- PMID: 38225249
- PMCID: PMC10789789
- DOI: 10.1038/s41467-023-44680-3
Large scale plasma proteomics identifies novel proteins and protein networks associated with heart failure development
Abstract
Heart failure (HF) causes substantial morbidity and mortality but its pathobiology is incompletely understood. The proteome is a promising intermediate phenotype for discovery of novel mechanisms. We measured 4877 plasma proteins in 13,900 HF-free individuals across three analysis sets with diverse age, geography, and HF ascertainment to identify circulating proteins and protein networks associated with HF development. Parallel analyses in Atherosclerosis Risk in Communities study participants in mid-life and late-life and in Trøndelag Health Study participants identified 37 proteins consistently associated with incident HF independent of traditional risk factors. Mendelian randomization supported causal effects of 10 on HF, HF risk factors, or left ventricular size and function, including matricellular (e.g. SPON1, MFAP4), senescence-associated (FSTL3, IGFBP7), and inflammatory (SVEP1, CCL15, ITIH3) proteins. Protein co-regulation network analyses identified 5 modules associated with HF risk, two of which were influenced by genetic variants that implicated trans hotspots within the VTN and CFH genes.
© 2024. The Author(s).
Conflict of interest statement
The Atherosclerosis Risk in Communities study has been funded in whole or in part with Federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, Department of Health and Human Services, under Contract nos. (HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700005I, HHSN268201700004I). Dr Shah was supported by NIH/NHLBI grants R01HL135008, R01HL143224, R01HL150342, R01HL148218, R01HL160025, and K24HL152008. Drs Myhre and Omland were supported by Research Council of Norway grant 296357/TOE. Dr Buckley was supported by NIH/NHLBI grant K23HL150311. Dr. Ballantyne was supported by NIH/NHLBI grant R01HL134320. Dr. Yu was in part supported by R01HL148218, R01 HL160793 and the JLH Foundation. The funder had no role design and conduct of this study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication. Dr. Shah reports research support from Novartis through Brigham and Women’s Hospital and consulting fees from Philips Ultrasound and Janssen. Dr. Myhre has consulted for and/or received speaker honoraria from Amarin, AmGen, Bayer, AstraZeneca, Boehringer-Ingelheim, Novartis, Novo Nordisk, Pharmacosmos, and Us2.ai. Dr Dalen holds positions at Centre for Innovative Ultrasound Solutions (CIUS) and Precision Health Center for optimized cardiac care (ProCardio) - both Norwegian Research Council (NRC) centers for research-based innovation, where GE Healthcare, Horten, Norway is one of the institutional partners, has research contracts with GE Healthcare, and acts as research advisor for Boehringer Ingelheim. Dr. Hoogeveen reports research support from Denka Seiken through Baylor College of Medicine and consulting fees from Denka Seiken. Dr Ballantyne reports grant/research support from Abbott Diagnostic, Akcea, Amgen, Arrowhead, Esperion, Ionis, Merck, Novartis, Novo Nordisk, Regeneron, Roche Diagnostic, NIH, AHA, and ADA, and is a consultant for 89Bio, Abbott Diagnostics, Alnylam Pharmaceuticals, Althera, Amarin, Amgen, Arrowhead, Astra Zeneca, Denka Seiken, Esperion, Genentech, Gilead, Illumina, Ionis, Matinas BioPharma Inc, Merck, New Amsterdam, Novartis, Novo Nordisk, Pfizer, Regeneron, Roche Diagnostic. Dr. Coresh is a scientific advisor to Soma Logic. The remaining authors report no competing interests.
Figures

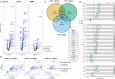



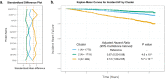

References
-
- Mozaffarian D, et al. Heart disease and stroke statistics-−2015 update: a report from the American Heart Association. Circulation. 2015;131:e29–e322. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

