Vaccine design via antigen reorientation
- PMID: 38225471
- PMCID: PMC11247139
- DOI: 10.1038/s41589-023-01529-6
Vaccine design via antigen reorientation
Abstract
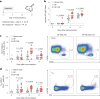
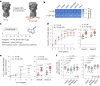
A major challenge in creating universal influenza vaccines is to focus immune responses away from the immunodominant, variable head region of hemagglutinin (HA-head) and toward the evolutionarily conserved stem region (HA-stem). Here we introduce an approach to control antigen orientation via site-specific insertion of aspartate residues that facilitates antigen binding to alum. We demonstrate the generalizability of this approach with antigens from Ebola, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and influenza viruses and observe enhanced neutralizing antibody responses in all cases. We then reorient an H2 HA in an 'upside-down' configuration to increase the exposure and immunogenicity of HA-stem. The reoriented H2 HA (reoH2HA) on alum induced stem-directed antibodies that cross-react with both group 1 and group 2 influenza A subtypes. Electron microscopy polyclonal epitope mapping (EMPEM) revealed that reoH2HA (group 1) elicits cross-reactive antibodies targeting group 2 HA-stems. Our results highlight antigen reorientation as a generalizable approach for designing epitope-focused vaccines.
© 2024. The Author(s).
Conflict of interest statement
D.X., P.A.B.W. and P.S.K. are named as inventors on a patent application applied for by Stanford University and the Chan Zuckerberg Biohub on engineering antigen binding and orientation on alum adjuvants. P.A.B.W. is an employee of Vaccine Company, Inc., and P.S.K. is a co-founder and member of the Board of Directors of Vaccine Company, Inc. All other authors declare no competing interests.
Figures










References
-
- Cox, N. J. & Subbarao, K. Global epidemiology of influenza: past and present. Annu. Rev. Med.51, 407–421 (2000). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

