Multipronged regulation of autophagy and apoptosis: emerging role of TRIM proteins
- PMID: 38225560
- PMCID: PMC10790450
- DOI: 10.1186/s11658-023-00528-8
Multipronged regulation of autophagy and apoptosis: emerging role of TRIM proteins
Abstract
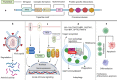
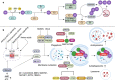
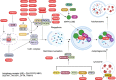
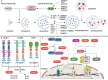
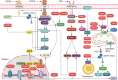
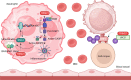
TRIM proteins are characterized by their conserved N-terminal RING, B-box, and coiled-coil domains. These proteins are efficient regulators of autophagy, apoptosis, and innate immune responses and confer immunity against viruses and bacteria. TRIMs function as receptors or scaffold proteins that target substrates for autophagy-mediated degradation. Most TRIMs interact with the BECN1-ULK1 complex to form TRIMosomes, thereby efficiently targeting substrates to autophagosomes. They regulate the functions of ATG proteins through physical interactions or ubiquitination. TRIMs affect the lipidation of MAP1LC3B1 to form MAP1LC3B2, which is a prerequisite for phagophore and autophagosome formation. In addition, they regulate MTOR kinase and TFEB, thereby regulating the expression of ATG genes. TRIM proteins are efficient regulators of apoptosis and are crucial for regulating cell proliferation and tumor formation. Many TRIM proteins regulate intrinsic and extrinsic apoptosis via the cell surface receptors TGFBR2, TNFRSF1A, and FAS. Mitochondria modulate the anti- and proapoptotic functions of BCL2, BAX, BAK1, and CYCS. These proteins use a multipronged approach to regulate the intrinsic and extrinsic apoptotic pathways, culminating in coordinated activation or inhibition of the initiator and executor CASPs. Furthermore, TRIMs can have a dual effect in determining cell fate and are therefore crucial for cellular homeostasis. In this review, we discuss mechanistic insights into the role of TRIM proteins in regulating autophagy and apoptosis, which can be used to better understand cellular physiology. These findings can be used to develop therapeutic interventions to prevent or treat multiple genetic and infectious diseases.
Keywords: Apoptosis; Autophagosome; Autophagy; Autophagy receptor; BECN1; E3-Ub ligase; TP53; TRIM proteins; ULK1; Ubiquitination.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

