Isotopic Depletion Increases the Spatial Resolution of FPOP Top-Down Mass Spectrometry Analysis
- PMID: 38226459
- PMCID: PMC10831798
- DOI: 10.1021/acs.analchem.3c03759
Isotopic Depletion Increases the Spatial Resolution of FPOP Top-Down Mass Spectrometry Analysis
Abstract
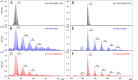
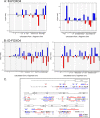
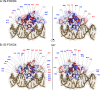
Protein radical labeling, like fast photochemical oxidation of proteins (FPOP), coupled to a top-down mass spectrometry (MS) analysis offers an alternative analytical method for probing protein structure or protein interaction with other biomolecules, for instance, proteins and DNA. However, with the increasing mass of studied analytes, the MS/MS spectra become complex and exhibit a low signal-to-noise ratio. Nevertheless, these difficulties may be overcome by protein isotope depletion. Thus, we aimed to use protein isotope depletion to analyze FPOP-oxidized samples by top-down MS analysis. For this purpose, we prepared isotopically natural (IN) and depleted (ID) forms of the FOXO4 DNA binding domain (FOXO4-DBD) and studied the protein-DNA interaction interface with double-stranded DNA, the insulin response element (IRE), after exposing the complex to hydroxyl radicals. As shown by comparing tandem mass spectra of natural and depleted proteins, the ID form increased the signal-to-noise ratio of useful fragment ions, thereby enhancing the sequence coverage by more than 19%. This improvement in the detection of fragment ions enabled us to detect 22 more oxidized residues in the ID samples than in the IN sample. Moreover, less common modifications were detected in the ID sample, including the formation of ketones and lysine carbonylation. Given the higher quality of ID top-down MSMS data set, these results provide more detailed information on the complex formation between transcription factors and DNA-response elements. Therefore, our study highlights the benefits of isotopic depletion for quantitative top-down proteomics. Data are available via ProteomeXchange with the identifier PXD044447.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
-
- Polák M.; Yassaghi G.; Kavan D.; Filandr F.; Fiala J.; Kukačka Z.; Halada P.; Loginov D. S.; Novák P. Utilization of Fast Photochemical Oxidation of Proteins and Both Bottom-up and Top-down Mass Spectrometry for Structural Characterization of a Transcription Factor–DsDNA Complex. Anal. Chem. 2022, 94 (7), 3203–3210. 10.1021/acs.analchem.1c04746. - DOI - PubMed
-
- Pandey P.; Hasnain S.; Ahmad S.. Protein-DNA Interactions. In Encyclopedia of Bioinformatics and Computational Biology; Elsevier, 2019; pp 142–154. 10.1016/B978-0-12-809633-8.20217-3. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

