Flexible active-site loops fine-tune substrate specificity of hyperthermophilic metallo-oxidases
- PMID: 38227199
- PMCID: PMC11111587
- DOI: 10.1007/s00775-023-02040-y
Flexible active-site loops fine-tune substrate specificity of hyperthermophilic metallo-oxidases
Abstract
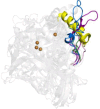
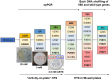
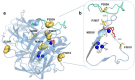
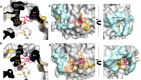
Hyperthermophilic ('superheat-loving') archaea found in high-temperature environments such as Pyrobaculum aerophilum contain multicopper oxidases (MCOs) with remarkable efficiency for oxidizing cuprous and ferrous ions. In this work, directed evolution was used to expand the substrate specificity of P. aerophilum McoP for organic substrates. Six rounds of error-prone PCR and DNA shuffling followed by high-throughput screening lead to the identification of a hit variant with a 220-fold increased efficiency (kcat/Km) than the wild-type for 2,2'-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) (ABTS) without compromising its intrinsic activity for metal ions. The analysis of the X-ray crystal structure reveals four proximal mutations close to the T1Cu active site. One of these mutations is within the 23-residues loop that occludes this site, a distinctive feature of prokaryotic MCOs. The increased flexibility of this loop results in an enlarged tunnel and one additional pocket that facilitates bulky substrate-enzyme interactions. These findings underscore the synergy between mutations that modulate the dynamics of the active-site loop enabling enhanced catalytic function. This study highlights the potential of targeting loops close to the T1Cu for engineering improvements suitable for biotechnological applications.
Keywords: Directed evolution; Enzyme engineering; Enzyme specificity; Laccases; Multicopper oxidases.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
-
- Lindley PF. Multi-copper oxidases. In: Bertini I, Sigel A, Sigel H, editors. Handbook on metalloproteins. New York: Marcel Dekker; 2001. pp. 763–811.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

