The extracellular matrix supports breast cancer cell growth under amino acid starvation by promoting tyrosine catabolism
- PMID: 38227562
- PMCID: PMC10791009
- DOI: 10.1371/journal.pbio.3002406
The extracellular matrix supports breast cancer cell growth under amino acid starvation by promoting tyrosine catabolism
Abstract
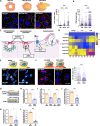
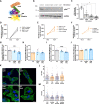
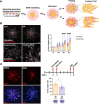
Breast tumours are embedded in a collagen I-rich extracellular matrix (ECM) network, where nutrients are scarce due to limited blood flow and elevated tumour growth. Metabolic adaptation is required for cancer cells to endure these conditions. Here, we demonstrated that the presence of ECM supported the growth of invasive breast cancer cells, but not non-transformed mammary epithelial cells, under amino acid starvation, through a mechanism that required macropinocytosis-dependent ECM uptake. Importantly, we showed that this behaviour was acquired during carcinoma progression. ECM internalisation, followed by lysosomal degradation, contributed to the up-regulation of the intracellular levels of several amino acids, most notably tyrosine and phenylalanine. This resulted in elevated tyrosine catabolism on ECM under starvation, leading to increased fumarate levels, potentially feeding into the tricarboxylic acid (TCA) cycle. Interestingly, this pathway was required for ECM-dependent cell growth and invasive cell migration under amino acid starvation, as the knockdown of p-hydroxyphenylpyruvate hydroxylase-like protein (HPDL), the third enzyme of the pathway, opposed cell growth and motility on ECM in both 2D and 3D systems, without affecting cell proliferation on plastic. Finally, high HPDL expression correlated with poor prognosis in breast cancer patients. Collectively, our results highlight that the ECM in the tumour microenvironment (TME) represents an alternative source of nutrients to support cancer cell growth by regulating phenylalanine and tyrosine metabolism.
Copyright: © 2024 Nazemi et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

