Exploring the resistance mechanism of triple-negative breast cancer to paclitaxel through the scRNA-seq analysis
- PMID: 38227591
- PMCID: PMC10791000
- DOI: 10.1371/journal.pone.0297260
Exploring the resistance mechanism of triple-negative breast cancer to paclitaxel through the scRNA-seq analysis
Abstract
Background: The triple negative breast cancer (TNBC) is the most malignant subtype of breast cancer with high aggressiveness. Although paclitaxel-based chemotherapy scenario present the mainstay in TNBC treatment, paclitaxel resistance is still a striking obstacle for cancer cure. So it is imperative to probe new therapeutic targets through illustrating the mechanisms underlying paclitaxel chemoresistance.
Methods: The Single cell RNA sequencing (scRNA-seq) data of TNBC cells treated with paclitaxel at different points were downloaded from the Gene Expression Omnibus (GEO) database. The Seurat R package was used to filter and integrate the scRNA-seq expression matrix. Cells were further clustered by the FindClusters function, and the gene marker of each subset was defined by FindAllMarkers function. Then, the hallmark score of each cell was calculated by AUCell R package, the biological function of the highly expressed interest genes was analyzed by the DAVID database. Subsequently, we performed pseudotime analysis to explore the change patterns of drug resistance genes and SCENIC analysis to identify the key transcription factors (TFs). Finally, the inhibitors of which were also analyzed by the CTD database.
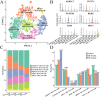
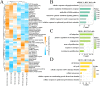
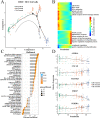
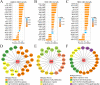
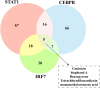
Results: We finally obtained 6 cell subsets from 2798 cells, which were marked as AKR1C3+, WNT7A+, FAM72B+, RERG+, IDO1+ and HEY1+HCC1143 cell subsets, among which the AKR1C3+, IDO1+ and HEY1+ cell subsets proportions increased with increasing treatment time, and then were regarded as paclitaxel resistance subsets. Hallmark score and pseudotime analysis showed that these paclitaxel resistance subsets were associated with the inflammatory response, virus and interferon response activation. In addition, the gene regulatory networks (GRNs) indicated that 3 key TFs (STAT1, CEBPB and IRF7) played vital role in promoting resistance development, and five common inhibitors targeted these TFs as potential combination therapies of paclitaxel were identified.
Conclusion: In this study, we identified 3 paclitaxel resistance relevant IFs and their inhibitors, which offers essential molecular basis for paclitaxel resistance and beneficial guidance for the combination of paclitaxel in clinical TNBC therapy.
Copyright: © 2024 Gao et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Revealing Cellular Heterogeneity and Key Regulatory Factors of Triple-Negative Breast Cancer through Single-Cell RNA Sequencing.Front Biosci (Landmark Ed). 2024 Aug 19;29(8):290. doi: 10.31083/j.fbl2908290. Front Biosci (Landmark Ed). 2024. PMID: 39206896
-
TNBC response to paclitaxel phenocopies interferon response which reveals cell cycle-associated resistance mechanisms.Sci Rep. 2025 Feb 4;15(1):4294. doi: 10.1038/s41598-024-82218-9. Sci Rep. 2025. PMID: 39905117 Free PMC article.
-
Identification of New Chemoresistance-Associated Genes in Triple-Negative Breast Cancer by Single-Cell Transcriptomic Analysis.Int J Mol Sci. 2024 Jun 22;25(13):6853. doi: 10.3390/ijms25136853. Int J Mol Sci. 2024. PMID: 38999963 Free PMC article.
-
SYTL4 downregulates microtubule stability and confers paclitaxel resistance in triple-negative breast cancer.Theranostics. 2020 Aug 29;10(24):10940-10956. doi: 10.7150/thno.45207. eCollection 2020. Theranostics. 2020. PMID: 33042263 Free PMC article.
-
Truncated HDAC9 identified by integrated genome-wide screen as the key modulator for paclitaxel resistance in triple-negative breast cancer.Theranostics. 2020 Sep 2;10(24):11092-11109. doi: 10.7150/thno.44997. eCollection 2020. Theranostics. 2020. PMID: 33042272 Free PMC article.
Cited by
-
Kang Ru enhances paclitaxel's efficacy against breast cancer progression.Am J Cancer Res. 2025 May 15;15(5):2180-2192. doi: 10.62347/XMGX2636. eCollection 2025. Am J Cancer Res. 2025. PMID: 40520866 Free PMC article.
-
Targeting Resistance Pathways in Breast Cancer Through Precision Oncology: Nanotechnology and Immune Modulation Approaches.Biomedicines. 2025 Jul 10;13(7):1691. doi: 10.3390/biomedicines13071691. Biomedicines. 2025. PMID: 40722763 Free PMC article. Review.
-
BUB1 Inhibition Sensitizes TNBC Cell Lines to Chemotherapy and Radiotherapy.Biomolecules. 2024 May 25;14(6):625. doi: 10.3390/biom14060625. Biomolecules. 2024. PMID: 38927028 Free PMC article.
-
Real world analysis of the efficacy and safety of eribulin compared to utidelone in combination with capecitabine for the treatment of metastatic breast cancer.Cancer Cell Int. 2024 Dec 19;24(1):416. doi: 10.1186/s12935-024-03608-7. Cancer Cell Int. 2024. PMID: 39702163 Free PMC article.
-
Inhibition of NLRP3 inflammasome contributes to paclitaxel efficacy in triple negative breast cancer treatment.Sci Rep. 2024 Oct 21;14(1):24753. doi: 10.1038/s41598-024-75805-3. Sci Rep. 2024. PMID: 39433537 Free PMC article.
References
-
- Shang C, Xu D. Epidemiology of Breast Cancer. Oncologie. 2022;24(4):649–63.
-
- Roulot A, Héquet D, Guinebretière JM, Vincent-Salomon A, Lerebours F, Dubot C, et al.. Tumoral heterogeneity of breast cancer. Ann Biol Clin (Paris). 2016;74(6):653–60. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

