TREX1 Inactivation Unleashes Cancer Cell STING-Interferon Signaling and Promotes Antitumor Immunity
- PMID: 38227896
- PMCID: PMC11062818
- DOI: 10.1158/2159-8290.CD-23-0700
TREX1 Inactivation Unleashes Cancer Cell STING-Interferon Signaling and Promotes Antitumor Immunity
Abstract
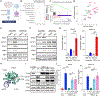
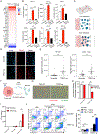
A substantial fraction of cancers evade immune detection by silencing Stimulator of Interferon Genes (STING)-Interferon (IFN) signaling. Therapeutic reactivation of this program via STING agonists, epigenetic, or DNA-damaging therapies can restore antitumor immunity in multiple preclinical models. Here we show that adaptive induction of three prime exonuclease 1 (TREX1) restrains STING-dependent nucleic acid sensing in cancer cells via its catalytic function in degrading cytosolic DNA. Cancer cell TREX1 expression is coordinately induced with STING by autocrine IFN and downstream STAT1, preventing signal amplification. TREX1 inactivation in cancer cells thus unleashes STING-IFN signaling, recruiting T and natural killer (NK) cells, sensitizing to NK cell-derived IFNγ, and cooperating with programmed cell death protein 1 blockade in multiple mouse tumor models to enhance immunogenicity. Targeting TREX1 may represent a complementary strategy to induce cytosolic DNA and amplify cancer cell STING-IFN signaling as a means to sensitize tumors to immune checkpoint blockade (ICB) and/or cell therapies.
Significance: STING-IFN signaling in cancer cells promotes tumor cell immunogenicity. Inactivation of the DNA exonuclease TREX1, which is adaptively upregulated to limit pathway activation in cancer cells, recruits immune effector cells and primes NK cell-mediated killing. Targeting TREX1 has substantial therapeutic potential to amplify cancer cell immunogenicity and overcome ICB resistance. This article is featured in Selected Articles from This Issue, p. 695.
©2024 American Association for Cancer Research.
Conflict of interest statement
Declaration of Interest
D.A.B. is a consultant for N of One/Qiagen and Nerviano Medical Sciences, is a founder and shareholder in Xsphera Biosciences, has received honoraria from Merck, H3 Biomedicine/Esai, EMD Serono, Gilead Sciences, Abbvie, and Madalon Consulting, and research grants from BMS, Takeda, Novartis, Gilead, and Lilly. C.P.P is a consultant for DropWorks and XSphera Biosciences, has stock and other ownership interests in XSphera Biosciences. Received honoraria from Bio-Rad and has sponsored research agreements with Daiichi Sankyo, Bicycle Therapeutics, Transcenta, Bicara Therapeutics, AstraZeneca, Intellia Therapeutics, Janssen Pharmaceuticals, Array Biopharma. S.K. has a sponsored research agreement with Boehringer-Ingelheim. H.M., A.K., J.P, M.W., B.J.W. and J.C. are employees of Gilead Sciences.
Figures


References
-
- Kitajima S, Ivanova E, Guo S, Yoshida R, Campisi M, Sundararaman SK, et al. Suppression of STING Associated with LKB1 Loss in KRAS-Driven Lung Cancer. Cancer Discov 2019;9(1):34–45 doi 10.1158/2159-8290.Cd-18-0689. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

