Selective CK1α degraders exert antiproliferative activity against a broad range of human cancer cell lines
- PMID: 38228616
- PMCID: PMC10791743
- DOI: 10.1038/s41467-024-44698-1
Selective CK1α degraders exert antiproliferative activity against a broad range of human cancer cell lines
Abstract
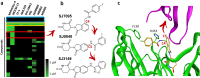
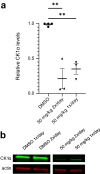
Molecular-glue degraders are small molecules that induce a specific interaction between an E3 ligase and a target protein, resulting in the target proteolysis. The discovery of molecular glue degraders currently relies mostly on screening approaches. Here, we describe screening of a library of cereblon (CRBN) ligands against a panel of patient-derived cancer cell lines, leading to the discovery of SJ7095, a potent degrader of CK1α, IKZF1 and IKZF3 proteins. Through a structure-informed exploration of structure activity relationship (SAR) around this small molecule we develop SJ3149, a selective and potent degrader of CK1α protein in vitro and in vivo. The structure of SJ3149 co-crystalized in complex with CK1α + CRBN + DDB1 provides a rationale for the improved degradation properties of this compound. In a panel of 115 cancer cell lines SJ3149 displays a broad antiproliferative activity profile, which shows statistically significant correlation with MDM2 inhibitor Nutlin-3a. These findings suggest potential utility of selective CK1α degraders for treatment of hematological cancers and solid tumors.
© 2024. The Author(s).
Conflict of interest statement
The authors declare the following competing interests: St. Jude Children’s Research Hospital has applied for an international patent covering structures reported in this work (WO2023081224A1; and provisional application number: 63389477), G.N., K.M., J.M.K., and Z.R. The remaining authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

