Osteocalcin of maternal and embryonic origins synergize to establish homeostasis in offspring
- PMID: 38228788
- PMCID: PMC10897216
- DOI: 10.1038/s44319-023-00031-3
Osteocalcin of maternal and embryonic origins synergize to establish homeostasis in offspring
Abstract
Many physiological osteocalcin-regulated functions are affected in adult offspring of mothers experiencing unhealthy pregnancy. Furthermore, osteocalcin signaling during gestation influences cognition and adrenal steroidogenesis in adult mice. Together these observations suggest that osteocalcin may broadly function during pregnancy to determine organismal homeostasis in adult mammals. To test this hypothesis, we analyzed in unchallenged wildtype and Osteocalcin-deficient, newborn and adult mice of various genotypes and origin maintained on different genetic backgrounds, the functions of osteocalcin in the pancreas, liver and testes and their molecular underpinnings. This analysis revealed that providing mothers are Osteocalcin-deficient, Osteocalcin haploinsufficiency in embryos hampers insulin secretion, liver gluconeogenesis, glucose homeostasis, testes steroidogenesis in adult offspring; inhibits cell proliferation in developing pancreatic islets and testes; and disrupts distinct programs of gene expression in these organs and in the brain. This study indicates that osteocalcin exerts dominant functions in most organs it influences. Furthermore, through their synergistic regulation of multiple physiological functions, osteocalcin of maternal and embryonic origins contributes to the establishment and maintenance of organismal homeostasis in newborn and adult offspring.
Keywords: Developmental Effect; Osteocalcin; Postnatal Physiology.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

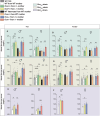
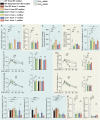
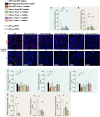

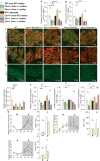




Update of
-
Osteocalcin of maternal and embryonic origins synergize to establish homeostasis in offspring.bioRxiv [Preprint]. 2023 Aug 14:2023.08.11.552969. doi: 10.1101/2023.08.11.552969. bioRxiv. 2023. Update in: EMBO Rep. 2024 Feb;25(2):593-615. doi: 10.1038/s44319-023-00031-3. PMID: 37645714 Free PMC article. Updated. Preprint.
References
-
- Accili D, Drago J, Lee EJ, Johnson MD, Cool MH, Salvatore P, Asico LD, Jose PA, Taylor SI, Westphal H. Early neonatal death in mice homozygous for a null allele of the insulin receptor gene. Nat Genet. 1996;12:106–109. - PubMed
-
- Ashworth CJ, Hoggard N, Thomas L, Mercer JG, Wallace JM, Lea RG. Placental leptin. Rev Reprod. 2000;5:18–24. - PubMed
-
- Burger G, Drummond J, Sandstead H (1948) Malnutrition and starvation in Western Netherlands. Part I. General State Printing Office, The Hague
-
- Chan YF, Wai-Sum O, Tang F. Adrenomedullin in the rat testis. I: Its production, actions on testosterone secretion, regulation by human chorionic gonadotropin, and its interaction with endothelin 1 in the leydig cell. Biol Reprod. 2008;78:773–779. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

