Intra-tumoral T cells in pediatric brain tumors display clonal expansion and effector properties
- PMID: 38228835
- PMCID: PMC12496457
- DOI: 10.1038/s43018-023-00706-9
Intra-tumoral T cells in pediatric brain tumors display clonal expansion and effector properties
Abstract
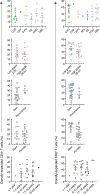
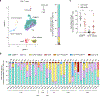
Brain tumors in children are a devastating disease in a high proportion of patients. Owing to inconsistent results in clinical trials in unstratified patients, the role of immunotherapy remains unclear. We performed an in-depth survey of the single-cell transcriptomes and clonal relationship of intra-tumoral T cells from children with brain tumors. Our results demonstrate that a large fraction of T cells in the tumor tissue are clonally expanded with the potential to recognize tumor antigens. Such clonally expanded T cells display enrichment of transcripts linked to effector function, tissue residency, immune checkpoints and signatures of neoantigen-specific T cells and immunotherapy response. We identify neoantigens in pediatric brain tumors and show that neoantigen-specific T cell gene signatures are linked to better survival outcomes. Notably, among the patients in our cohort, we observe substantial heterogeneity in the degree of clonal expansion and magnitude of T cell response. Our findings suggest that characterization of intra-tumoral T cell responses may enable selection of patients for immunotherapy, an approach that requires prospective validation in clinical trials.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing Interests
The authors declare no competing financial and / or non-financial interests.
Figures














References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

