NUSAP1 promotes pancreatic ductal adenocarcinoma progression by drives the epithelial-mesenchymal transition and reduces AMPK phosphorylation
- PMID: 38229038
- PMCID: PMC10790387
- DOI: 10.1186/s12885-024-11842-5
NUSAP1 promotes pancreatic ductal adenocarcinoma progression by drives the epithelial-mesenchymal transition and reduces AMPK phosphorylation
Abstract
Background: Pancreatic ductal adenocarcinoma (PDAC) has a poor prognosis, and its molecular mechanisms are unclear. Nucleolar and spindle-associated protein 1 (NUSAP1), an indispensable mitotic regulator, has been reported to be involved in the development of several types of tumors. The biological function and molecular mechanism of NUSAP1 in PDAC remain controversial. This study explored the effects and mechanism of NUSAP1 in PDAC.
Methods: Differentially expressed genes (DEGs) were screened. A protein‒protein interaction (PPI) network was constructed to identify hub genes. Experimental studies and tissue microarray (TMA) analysis were performed to investigate the effects of NUSAP1 in PDAC and explore its mechanism.
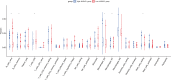
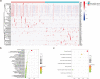
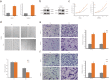
Results: Network analysis revealed that NUSAP1 is an essential hub gene in the PDAC transcriptome. Genome heterogeneity analysis revealed that NUSAP1 is related to tumor mutation burden (TMB), loss of heterozygosity (LOH) and homologous recombination deficiency (HRD) in PDAC. NUSAP1 is correlated with the levels of infiltrating immune cells, such as B cells and CD8 T cells. High NUSAP1 expression was found in PDAC tissues and was associated with a poor patient prognosis. NUSAP1 promoted cancer cell proliferation, migration and invasion, drives the epithelial-mesenchymal transition and reduces AMPK phosphorylation.
Conclusions: NUSAP1 is an essential hub gene that promotes PDAC progression and leads to a dismal prognosis by drives the epithelial-mesenchymal transition and reduces AMPK phosphorylation.
Keywords: Differentially expressed genes; Functional enrichment analysis; Pancreatic ductal adenocarcinoma; Protein‒protein interaction; Survival analysis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
-
- Huang H, Wright S, Zhang J, Brekken RA. Getting a grip on adhesion: cadherin switching and collagen signaling. Biochim Biophys Acta Mol Cell Res. 2019;1866(11):118472. - PubMed
-
- Strobel O, Neoptolemos J, Jäger D, Büchler MW. Optimizing the outcomes of pancreatic cancer surgery. Nat Rev Clin Oncol. 2019;16(1):11–26. - PubMed
MeSH terms
Substances
Grants and funding
- U21A20374/the National Natural Science Foundation of China
- U21A20374/the National Natural Science Foundation of China
- U21A20374/the National Natural Science Foundation of China
- U21A20374/the National Natural Science Foundation of China
- U21A20374/the National Natural Science Foundation of China
- U21A20374/the National Natural Science Foundation of China
- U21A20374/the National Natural Science Foundation of China
- U21A20374/the National Natural Science Foundation of China
- 21JC1401500/Shanghai Municipal Science and Technology Major Project
- 21JC1401500/Shanghai Municipal Science and Technology Major Project
- 21JC1401500/Shanghai Municipal Science and Technology Major Project
- 21JC1401500/Shanghai Municipal Science and Technology Major Project
- 21JC1401500/Shanghai Municipal Science and Technology Major Project
- 21JC1401500/Shanghai Municipal Science and Technology Major Project
- 21JC1401500/Shanghai Municipal Science and Technology Major Project
- 21JC1401500/Shanghai Municipal Science and Technology Major Project
- 2019-01-07-00-07-E00057/Scientific Innovation Project of Shanghai Education Committee
- 2019-01-07-00-07-E00057/Scientific Innovation Project of Shanghai Education Committee
- 2019-01-07-00-07-E00057/Scientific Innovation Project of Shanghai Education Committee
- 2019-01-07-00-07-E00057/Scientific Innovation Project of Shanghai Education Committee
- 2019-01-07-00-07-E00057/Scientific Innovation Project of Shanghai Education Committee
- 2019-01-07-00-07-E00057/Scientific Innovation Project of Shanghai Education Committee
- 2019-01-07-00-07-E00057/Scientific Innovation Project of Shanghai Education Committee
- 2019-01-07-00-07-E00057/Scientific Innovation Project of Shanghai Education Committee
- SHDC2020CR1006A/Clinical Research Plan of Shanghai Hospital Development Center
- SHDC2020CR1006A/Clinical Research Plan of Shanghai Hospital Development Center
- SHDC2020CR1006A/Clinical Research Plan of Shanghai Hospital Development Center
- SHDC2020CR1006A/Clinical Research Plan of Shanghai Hospital Development Center
- SHDC2020CR1006A/Clinical Research Plan of Shanghai Hospital Development Center
- SHDC2020CR1006A/Clinical Research Plan of Shanghai Hospital Development Center
- SHDC2020CR1006A/Clinical Research Plan of Shanghai Hospital Development Center
- SHDC2020CR1006A/Clinical Research Plan of Shanghai Hospital Development Center
- 2021-011/Xuhui District Artificial Intelligence Medical Hospital Cooperation Project
- 2021-011/Xuhui District Artificial Intelligence Medical Hospital Cooperation Project
- 2021-011/Xuhui District Artificial Intelligence Medical Hospital Cooperation Project
- 2021-011/Xuhui District Artificial Intelligence Medical Hospital Cooperation Project
- 2021-011/Xuhui District Artificial Intelligence Medical Hospital Cooperation Project
- 2021-011/Xuhui District Artificial Intelligence Medical Hospital Cooperation Project
- 2021-011/Xuhui District Artificial Intelligence Medical Hospital Cooperation Project
- 2021-011/Xuhui District Artificial Intelligence Medical Hospital Cooperation Project
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

