Characterization of the complete chloroplast genome of Gastrochilus sinensis (Orchidaceae, Epidendroideae), a beautiful epiphytic orchid from China
- PMID: 38229739
- PMCID: PMC10791082
- DOI: 10.1080/23802359.2023.2301033
Characterization of the complete chloroplast genome of Gastrochilus sinensis (Orchidaceae, Epidendroideae), a beautiful epiphytic orchid from China
Abstract
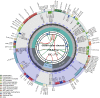
Gastrochilus sinensis is a beautiful epiphytic orchid with high ornamental value. In this study, the first complete chloroplast genome sequence of G. sinensis was determined using next-generation sequencing (NGS). The de novo assembled chloroplast genome was 148,020 bp in length, including a pair of inverted repeat regions (IRs; 25,987 bp), a small single-copy region (SSC; 11,045 bp), and a large single-copy region (LSC; 85,001 bp). The chloroplast genome encodes 109 unique genes, including 75 protein-coding genes (PCGs), 30 tRNA genes, and four rRNA genes. The total GC content of the chloroplast genome was 36.8%. The phylogenetic analysis showed a close relationship between G. sinensis and G. formosanus species. The complete chloroplast genome provides fundamental information for genetic diversity and phylogenetic relationships in Gastrochilus.
Keywords: Gastrochilus sinensis; chloroplast genome; phylogeny.
© 2024 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


References
-
- Chen SC, Liu ZJ, Zhu GH, Lang KY, Tsi ZH, Lou YB, Jin XH, Cribb PJ, Wood JJ, Gale SW, et al. . 2009. Orchidaceae. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 25. Beijing: Science Press; p. 497.
-
- Cheng YH, Chen YK, Ma J, He T, Zhang JY, Xu B.. 2022. Gastrochilus sinensis, a new record of Orchidaceae species in Sichuan Province, China. J Sichuan For Sci Technol. 43:139–142. doi:10.12172/202201250002. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
