Segregation of nascent GPCRs in the ER-to-Golgi transport by CCHCR1 via direct interaction
- PMID: 38230433
- PMCID: PMC10912811
- DOI: 10.1242/jcs.261685
Segregation of nascent GPCRs in the ER-to-Golgi transport by CCHCR1 via direct interaction
Abstract
G protein-coupled receptors (GPCRs) constitute the largest superfamily of cell surface signaling proteins that share a common structural topology. When compared with agonist-induced internalization, how GPCRs are sorted and delivered to functional destinations after synthesis in the endoplasmic reticulum (ER) is much less well understood. Here, we demonstrate that depletion of coiled-coil α-helical rod protein 1 (CCHCR1) by siRNA and CRISPR-Cas9 significantly inhibits surface expression and signaling of α2A-adrenergic receptor (α2A-AR; also known as ADRA2A), without affecting α2B-AR. Further studies show that CCHCR1 depletion specifically impedes α2A-AR export from the ER to the Golgi, but not from the Golgi to the surface. We also demonstrate that CCHCR1 selectively interacts with α2A-AR. The interaction is mediated through multiple domains of both proteins and is ionic in nature. Moreover, mutating CCHCR1-binding motifs significantly attenuates ER-to-Golgi export, surface expression and signaling of α2A-AR. Collectively, these data reveal a novel function for CCHCR1 in intracellular protein trafficking, indicate that closely related GPCRs can be sorted into distinct ER-to-Golgi transport routes by CCHCR1 via direct interaction, and provide important insights into segregation and anterograde delivery of nascent GPCR members.
Keywords: Adrenergic receptor; Biosynthesis; CCHCR1; ER export; G protein-coupled receptor; Sorting; Trafficking.
© 2024. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures
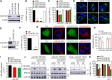

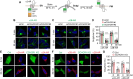
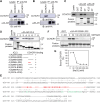



Similar articles
-
Human C1orf27 protein interacts with α2A-adrenergic receptor and regulates its anterograde transport.J Biol Chem. 2022 Jun;298(6):102021. doi: 10.1016/j.jbc.2022.102021. Epub 2022 May 10. J Biol Chem. 2022. PMID: 35551911 Free PMC article.
-
The GTPase Rab43 Controls the Anterograde ER-Golgi Trafficking and Sorting of GPCRs.Cell Rep. 2017 Oct 24;21(4):1089-1101. doi: 10.1016/j.celrep.2017.10.011. Cell Rep. 2017. PMID: 29069590 Free PMC article.
-
Specific TBC Domain-Containing Proteins Control the ER-Golgi-Plasma Membrane Trafficking of GPCRs.Cell Rep. 2019 Jul 9;28(2):554-566.e4. doi: 10.1016/j.celrep.2019.05.033. Cell Rep. 2019. PMID: 31291588 Free PMC article.
-
Regulation of G protein-coupled receptor export trafficking.Biochim Biophys Acta. 2007 Apr;1768(4):853-70. doi: 10.1016/j.bbamem.2006.09.008. Epub 2006 Sep 23. Biochim Biophys Acta. 2007. PMID: 17074298 Free PMC article. Review.
-
Regulation of α2B-Adrenerigc Receptor Export Trafficking by Specific Motifs.Prog Mol Biol Transl Sci. 2015;132:227-44. doi: 10.1016/bs.pmbts.2015.03.004. Epub 2015 Mar 31. Prog Mol Biol Transl Sci. 2015. PMID: 26055061 Free PMC article. Review.
Cited by
-
The ufmylation cascade controls COPII recruitment, anterograde transport, and sorting of nascent GPCRs at ER.Sci Adv. 2024 Jun 21;10(25):eadm9216. doi: 10.1126/sciadv.adm9216. Epub 2024 Jun 21. Sci Adv. 2024. PMID: 38905340 Free PMC article.
References
-
- Asumalahti, K., Laitinen, T., Itkonen-Vatjus, R., Lokki, M. L., Suomela, S., Snellman, E., Saarialho-Kere, U. and Kere, J. (2000). A candidate gene for psoriasis near HLA-C, HCR (Pg8), is highly polymorphic with a disease-associated susceptibility allele. Hum. Mol. Genet. 9, 1533-1542. 10.1093/hmg/9.10.1533 - DOI - PubMed
-
- Asumalahti, K., Veal, C., Laitinen, T., Suomela, S., Allen, M., Elomaa, O., Moser, M., de Cid, R., Ripatti, S. and Vorechovsky, I. (2002). Coding haplotype analysis supports HCR as the putative susceptibility gene for psoriasis at the MHC PSORS1 locus. Hum. Mol. Genet. 11, 589-597. 10.1093/hmg/11.5.589 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

