A new experimental evidence-weighted signaling pathway resource in FlyBase
- PMID: 38230566
- PMCID: PMC10911275
- DOI: 10.1242/dev.202255
A new experimental evidence-weighted signaling pathway resource in FlyBase
Abstract
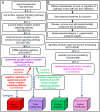
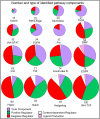
Research in model organisms is central to the characterization of signaling pathways in multicellular organisms. Here, we present the comprehensive and systematic curation of 17 Drosophila signaling pathways using the Gene Ontology framework to establish a dynamic resource that has been incorporated into FlyBase, providing visualization and data integration tools to aid research projects. By restricting to experimental evidence reported in the research literature and quantifying the amount of such evidence for each gene in a pathway, we captured the landscape of empirical knowledge of signaling pathways in Drosophila.
Keywords: Drosophila; Biocuration; FlyBase; Gene Ontology; Network; Signaling pathways.
© 2024. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures


Update of
-
A new experimental evidence-weighted signaling pathway resource in FlyBase.bioRxiv [Preprint]. 2023 Sep 15:2023.08.10.552786. doi: 10.1101/2023.08.10.552786. bioRxiv. 2023. Update in: Development. 2024 Feb 1;151(3):dev202255. doi: 10.1242/dev.202255. PMID: 37645956 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

