Nervous necrosis virus induced vacuolization is a Rab5- and actin-dependent process
- PMID: 38230744
- PMCID: PMC10795790
- DOI: 10.1080/21505594.2023.2301244
Nervous necrosis virus induced vacuolization is a Rab5- and actin-dependent process
Abstract
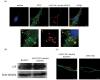
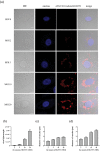
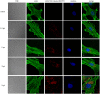
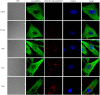
Cytoplasmic vacuolization is commonly induced by bacteria and viruses, reflecting the complex interactions between pathogens and the host. However, their characteristics and formation remain unclear. Nervous necrosis virus (NNV) infects more than 100 global fish species, causing enormous economic losses. Vacuolization is a hallmark of NNV infection in host cells, but remains a mystery. In this study, we developed a simple aptamer labelling technique to identify red-spotted grouper NNV (RGNNV) particles in fixed and live cells to explore RGNNV-induced vacuolization. We observed that RGNNV-induced vacuolization was positively associated with the infection time and virus uptake. During infection, most RGNNV particles, as well as viral genes, colocalized with vacuoles, but not giant vacuoles > 3 μm in diameter. Although the capsid protein (CP) is the only structural protein of RGNNV, its overexpression did not induce vacuolization, suggesting that vacuole formation probably requires virus entry and replication. Given that small Rab proteins and the cytoskeleton are key factors in regulating cellular vesicles, we further investigated their roles in RGNNV-induced vacuolization. Using live cell imaging, Rab5, a marker of early endosomes, was continuously located in vacuoles bearing RGNNV during giant vacuole formation. Rab5 is required for vacuole formation and interacts with CP according to siRNA interference and Co-IP analysis. Furthermore, actin formed distinct rings around small vacuoles, while vacuoles were located near microtubules. Actin, but not microtubules, plays an important role in vacuole formation using chemical inhibitors. These results provide valuable insights into the pathogenesis and control of RGNNV infections.
Keywords: RGNNV; Rab5; aptamer; imaging; vacuolization.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures





Similar articles
-
Nectin1 is a pivotal host factor involved in attachment and entry of red-spotted grouper nervous necrosis virus in the early stages of the viral life cycle.J Virol. 2024 Sep 17;98(9):e0090124. doi: 10.1128/jvi.00901-24. Epub 2024 Aug 28. J Virol. 2024. PMID: 39194240 Free PMC article.
-
Autophagy Participates in Lysosomal Vacuolation-Mediated Cell Death in RGNNV-Infected Cells.Front Microbiol. 2020 Apr 30;11:790. doi: 10.3389/fmicb.2020.00790. eCollection 2020. Front Microbiol. 2020. PMID: 32425913 Free PMC article.
-
Comparative transcriptomic analysis reveals different host cell responses to Singapore grouper iridovirus and red-spotted grouper nervous necrosis virus.Fish Shellfish Immunol. 2022 Sep;128:136-147. doi: 10.1016/j.fsi.2022.07.068. Epub 2022 Jul 31. Fish Shellfish Immunol. 2022. PMID: 35921938
-
Capsid protein from red-spotted grouper nervous necrosis virus induces incomplete autophagy by inactivating the HSP90ab1-AKT-MTOR pathway.Zool Res. 2022 Jan 18;43(1):98-110. doi: 10.24272/j.issn.2095-8137.2021.249. Zool Res. 2022. PMID: 34904422 Free PMC article.
-
Characterization of microRNAs in orange-spotted grouper (Epinephelus coioides) fin cells upon red-spotted grouper nervous necrosis virus infection.Fish Shellfish Immunol. 2017 Apr;63:228-236. doi: 10.1016/j.fsi.2017.02.031. Epub 2017 Feb 20. Fish Shellfish Immunol. 2017. PMID: 28232192
Cited by
-
Nectin1 is a pivotal host factor involved in attachment and entry of red-spotted grouper nervous necrosis virus in the early stages of the viral life cycle.J Virol. 2024 Sep 17;98(9):e0090124. doi: 10.1128/jvi.00901-24. Epub 2024 Aug 28. J Virol. 2024. PMID: 39194240 Free PMC article.
-
Rationally designed self-assembled peptide nanofibers provoke robust humoral immunity against nervous necrosis virus.J Virol. 2025 Aug 19;99(8):e0031925. doi: 10.1128/jvi.00319-25. Epub 2025 Jul 15. J Virol. 2025. PMID: 40662756 Free PMC article.
-
Fish Snx27 promotes viral products by modulating the innate immune response and exosomal machinery.J Virol. 2024 Dec 17;98(12):e0097424. doi: 10.1128/jvi.00974-24. Epub 2024 Nov 4. J Virol. 2024. PMID: 39494909 Free PMC article.
-
Proteomic and phosphoproteomic profilings reveal distinct cellular responses during Tilapinevirus tilapiae entry and replication.PeerJ. 2025 Feb 21;13:e18923. doi: 10.7717/peerj.18923. eCollection 2025. PeerJ. 2025. PMID: 39995988 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
