Preclinical Evaluation of NTX-301, a Novel DNA Hypomethylating Agent in Ovarian Cancer
- PMID: 38231483
- PMCID: PMC10947827
- DOI: 10.1158/1078-0432.CCR-23-2368
Preclinical Evaluation of NTX-301, a Novel DNA Hypomethylating Agent in Ovarian Cancer
Abstract
Purpose: DNA methylation causes silencing of tumor-suppressor and differentiation-associated genes, being linked to chemoresistance. Previous studies demonstrated that hypomethylating agents (HMA) resensitize ovarian cancer to chemotherapy. NTX-301 is a highly potent and orally bioavailable HMA, in early clinical development.
Experimental design: The antitumor effects of NTX-301 were studied in ovarian cancer models by using cell viability, stemness and ferroptosis assays, RNA sequencing, lipidomic analyses, and stimulated Raman spectroscopy.
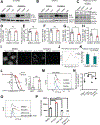
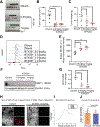
Results: Ovarian cancer cells (SKOV3, IC50 = 5.08 nmol/L; OVCAR5 IC50 = 3.66 nmol/L) were highly sensitive to NTX-301 compared with fallopian tube epithelial cells. NTX-301 downregulated expression of DNA methyltransferases 1-3 and induced transcriptomic reprogramming with 15,000 differentially expressed genes (DEG, P < 0.05). Among them, Gene Ontology enrichment analysis identified regulation of fatty acid biosynthesis and molecular functions related to aldehyde dehydrogenase (ALDH) and oxidoreductase, known features of cancer stem cells. Low-dose NTX-301 reduced the ALDH(+) cell population and expression of stemness-associated transcription factors. Stearoyl-coenzyme A desaturase 1 (SCD), which regulates production of unsaturated fatty acids (UFA), was among the top DEG downregulated by NTX-301. NTX-301 treatment decreased levels of UFA and increased oxidized lipids, and this was blunted by deferoxamine, indicating cell death via ferroptosis. NTX-301-induced ferroptosis was rescued by oleic acid. In vivo, monotherapy with NTX-301 significantly inhibited ovarian cancer and patient-derived xenograft growth (P < 0.05). Decreased SCD levels and increased oxidized lipids were detected in NTX-301-treated xenografts.
Conclusions: NTX-301 is active in ovarian cancer models. Our findings point to a new mechanism by which epigenetic blockade disrupts lipid homeostasis and promotes cancer cell death.
©2024 American Association for Cancer Research.
Conflict of interest statement
Conflict of Interest: PinotBio provided drug and funding to support part of the study.
Figures




Similar articles
-
CircASH1L inhibits ferroptosis and enhances cisplatin resistance by sponging miR-515-5p to regulate cell cycle-related CDCA7/RRM2 in ovarian cancer cells.Front Pharmacol. 2025 Jun 24;16:1563869. doi: 10.3389/fphar.2025.1563869. eCollection 2025. Front Pharmacol. 2025. PMID: 40630134 Free PMC article.
-
Impact of residual disease as a prognostic factor for survival in women with advanced epithelial ovarian cancer after primary surgery.Cochrane Database Syst Rev. 2022 Sep 26;9(9):CD015048. doi: 10.1002/14651858.CD015048.pub2. Cochrane Database Syst Rev. 2022. PMID: 36161421 Free PMC article.
-
Topotecan, pegylated liposomal doxorubicin hydrochloride and paclitaxel for second-line or subsequent treatment of advanced ovarian cancer: a systematic review and economic evaluation.Health Technol Assess. 2006 Mar;10(9):1-132. iii-iv. doi: 10.3310/hta10090. Health Technol Assess. 2006. PMID: 16545208
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of topotecan for ovarian cancer.Health Technol Assess. 2001;5(28):1-110. doi: 10.3310/hta5280. Health Technol Assess. 2001. PMID: 11701100
-
Pegylated liposomal doxorubicin for relapsed epithelial ovarian cancer.Cochrane Database Syst Rev. 2013 Jul 9;2013(7):CD006910. doi: 10.1002/14651858.CD006910.pub2. Cochrane Database Syst Rev. 2013. Update in: Cochrane Database Syst Rev. 2023 Jul 5;7:CD006910. doi: 10.1002/14651858.CD006910.pub3. PMID: 23835762 Free PMC article. Updated.
Cited by
-
Chromatin Organization Governs Transcriptional Response and Plasticity of Cancer Stem Cells.Adv Sci (Weinh). 2025 May;12(17):e2407426. doi: 10.1002/advs.202407426. Epub 2025 Mar 7. Adv Sci (Weinh). 2025. PMID: 40051293 Free PMC article.
-
Targeting ferroptosis: a promising avenue for ovarian cancer treatment.Front Immunol. 2025 Jun 5;16:1578723. doi: 10.3389/fimmu.2025.1578723. eCollection 2025. Front Immunol. 2025. PMID: 40539051 Free PMC article. Review.
-
First Indonesian Nasopharyngeal Cancer Whole Epigenome Sequencing Identify Tumour Suppressor CpG Methylation.Biologics. 2025 Jan 6;19:1-13. doi: 10.2147/BTT.S490382. eCollection 2025. Biologics. 2025. PMID: 39802100 Free PMC article.
References
-
- Kantarjian H, Issa JP, Rosenfeld CS, Bennett JM, Albitar M, DiPersio J, et al. Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer. 2006;106(8):1794–803. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

